Weight Distributions: w₁, ρ, and the Dual-Anchor Framework
JoonHo Lee
2026-01-01
Source:vignettes/theory-weights.Rmd
theory-weights.RmdOverview
This vignette provides a rigorous treatment of the stick-breaking weight distributions in Dirichlet Process (DP) models and their role in the dual-anchor elicitation framework. It is intended for statisticians and methodological researchers who wish to understand:
- The distributional properties of the first stick-breaking weight
- The co-clustering probability and its interpretation
- The functional for computing closed-form moments under Gamma hyperpriors
- The mathematical foundation of dual-anchor optimization
Throughout, we carefully distinguish between established results from the Bayesian nonparametrics literature and novel contributions of this work (the DPprior package and associated research notes).
Attribution Summary
| Result | Attribution |
|---|---|
| Stick-breaking construction | Sethuraman (1994) |
| GEM distribution / size-biased ordering | Kingman (1975), Pitman (1996), Arratia et al. (2003) |
| Kingman (1975), Pitman (1996) | |
| Closed-form distribution under Gamma prior | Vicentini & Jermyn (2025) |
| Dual-anchor framework and functional formulas | This work (DPprior package) |
1. Stick-Breaking Weights: Review and Interpretation
1.1 The GEM() Construction
Under Sethuraman’s (1994) stick-breaking representation of the DP, a random probability measure can be written as: where are the atom locations and are the stick-breaking weights constructed as:
The sequence follows the GEM() distribution (Griffiths-Engen-McCloskey), which represents a size-biased random permutation of the Poisson-Dirichlet distribution (Pitman, 1996).
1.2 Critical Interpretability Caveat
The GEM order is size-biased, NOT decreasing.
This distinction is essential for applied elicitation. Specifically:
- where the latter denotes the ranked (decreasing) weights
- is not “the largest cluster proportion”
- A faithful interpretation: is the asymptotic proportion of the cluster containing a randomly selected unit (equivalently, a size-biased pick from the Poisson-Dirichlet distribution)
Despite this caveat, remains a meaningful diagnostic for “dominance risk.” If is high, then a randomly selected unit is likely to belong to a cluster that contains more than half of the population.
# Demonstrate the stick-breaking construction
n_atoms <- 15
alpha_values <- c(0.5, 1, 2, 5)
set.seed(123)
sb_data <- do.call(rbind, lapply(alpha_values, function(a) {
v <- rbeta(n_atoms, 1, a)
w <- numeric(n_atoms)
w[1] <- v[1]
for (h in 2:n_atoms) {
w[h] <- v[h] * prod(1 - v[1:(h-1)])
}
data.frame(
atom = 1:n_atoms,
weight = w,
alpha = paste0("α = ", a)
)
}))
sb_data$alpha <- factor(sb_data$alpha, levels = paste0("α = ", alpha_values))
ggplot(sb_data, aes(x = factor(atom), y = weight, fill = alpha)) +
geom_bar(stat = "identity") +
facet_wrap(~alpha, nrow = 1) +
scale_fill_manual(values = palette_main) +
labs(
x = "Atom Index (h)",
y = expression(w[h]),
title = "Stick-Breaking Weights in GEM Order",
subtitle = "Single realization per α; NOT ranked by size"
) +
theme_minimal() +
theme(legend.position = "none")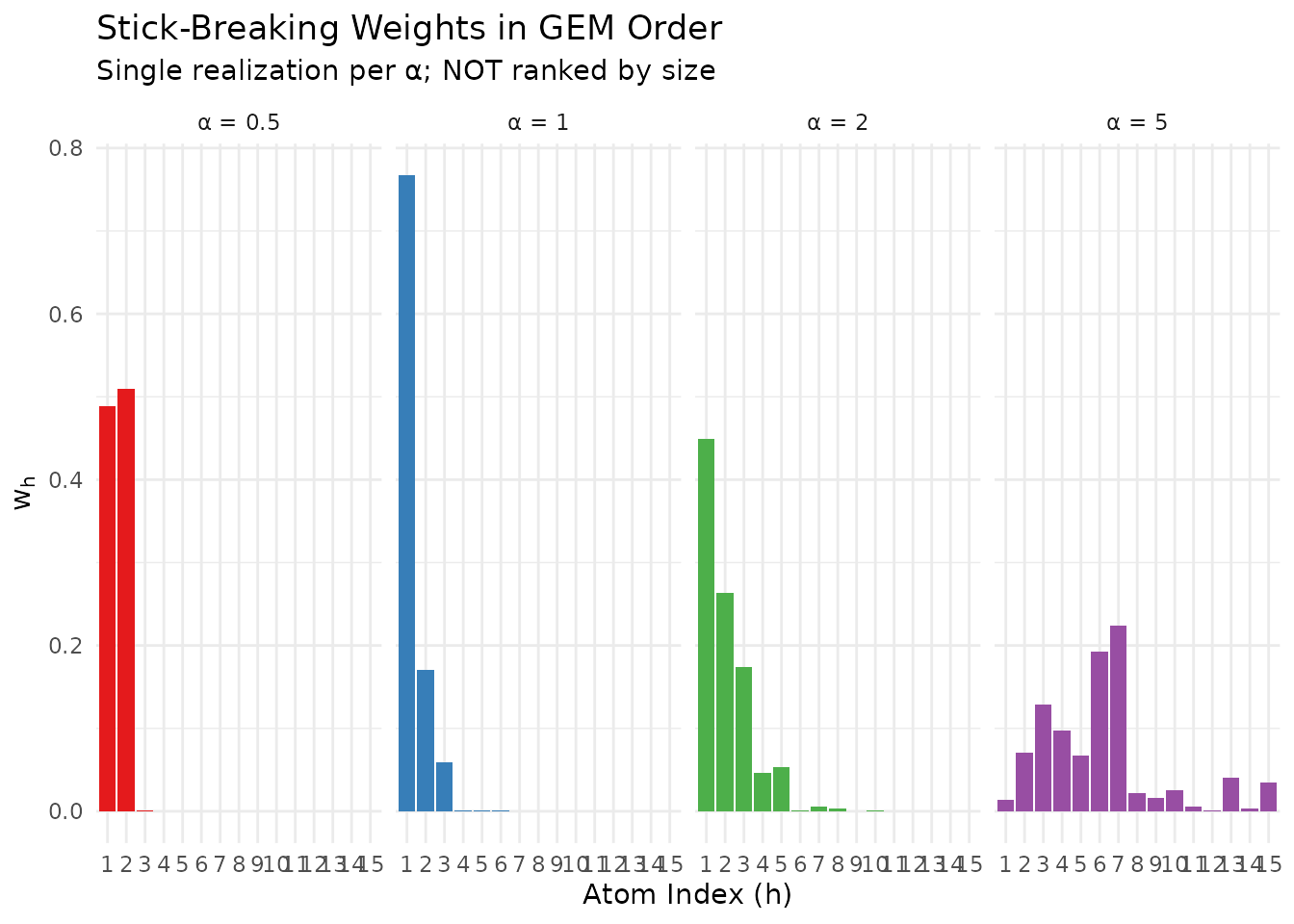
Stick-breaking weights for different α values. Smaller α concentrates mass on early atoms.
2. Distribution of
2.1 Conditional Distribution
The first stick-breaking weight has a particularly simple conditional distribution, following directly from the construction.
Proposition 1 (Conditional distribution of ). For :
Consequently:
- for
Attribution: This follows immediately from the stick-breaking construction (Sethuraman, 1994).
x_grid <- seq(0.001, 0.999, length.out = 200)
alpha_grid <- c(0.5, 1, 2, 5, 10)
cond_df <- do.call(rbind, lapply(alpha_grid, function(a) {
data.frame(
x = x_grid,
density = dbeta(x_grid, 1, a),
alpha = paste0("α = ", a)
)
}))
cond_df$alpha <- factor(cond_df$alpha, levels = paste0("α = ", alpha_grid))
ggplot(cond_df, aes(x = x, y = density, color = alpha)) +
geom_line(linewidth = 0.9) +
scale_color_viridis_d(option = "plasma", end = 0.85) +
labs(
x = expression(w[1]),
y = "Density",
title = expression("Conditional Density: " * w[1] * " | " * alpha * " ~ Beta(1, " * alpha * ")"),
color = NULL
) +
coord_cartesian(ylim = c(0, 6)) +
theme_minimal() +
theme(legend.position = "right")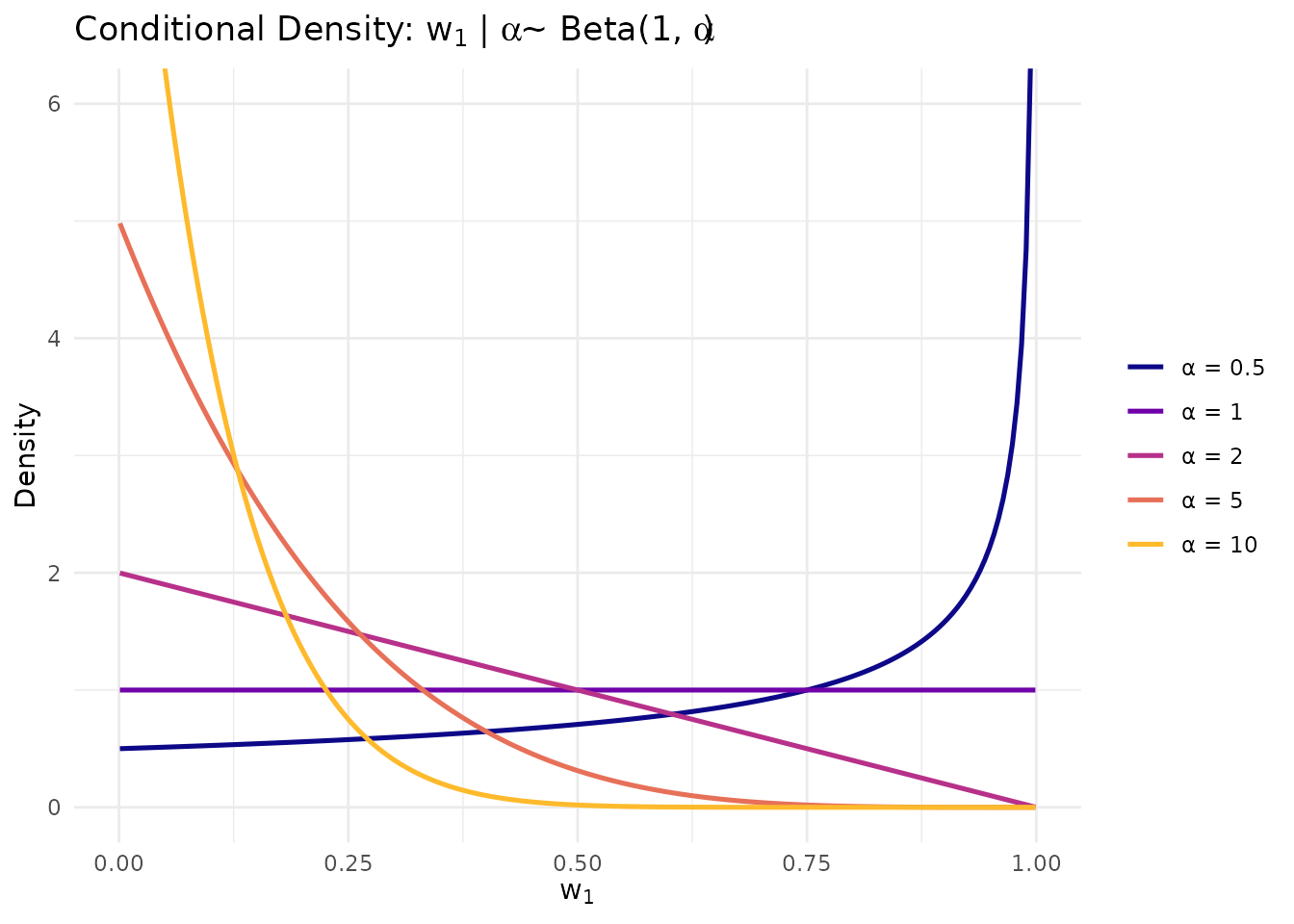
Conditional density of w₁|α for different concentration parameter values.
2.2 Marginal Distribution Under Gamma Hyperprior
When with shape and rate , the unconditional distribution of admits fully closed-form expressions.
Theorem 1 (Unconditional distribution of ; Vicentini & Jermyn, 2025, Appendix A). Let . Then:
Density:
CDF:
Survival function (dominance risk):
Quantile function:
Proof sketch. Integrate out from the conditional distribution: The integral evaluates to a Gamma function, and the CDF follows by direct integration with substitution .
Computational significance. These closed-form expressions enable computation of quantiles and tail probabilities—no Monte Carlo sampling is required.
# Demonstrate the closed-form w1 distribution functions
a <- 1.6
b <- 1.22 # The Lee et al. DP-inform prior
cat("w₁ distribution under Gamma(a=1.6, b=1.22) hyperprior:\n")
#> w₁ distribution under Gamma(a=1.6, b=1.22) hyperprior:
cat(sprintf(" Mean: %.4f\n", mean_w1(a, b)))
#> Mean: 0.5084
cat(sprintf(" Variance: %.4f\n", var_w1(a, b)))
#> Variance: 0.1052
cat(sprintf(" Median: %.4f\n", quantile_w1(0.5, a, b)))
#> Median: 0.4839
cat(sprintf(" 90th %%: %.4f\n", quantile_w1(0.9, a, b)))
#> 90th %: 0.9803
cat("\nDominance risk (tail probabilities):\n")
#>
#> Dominance risk (tail probabilities):
for (t in c(0.3, 0.5, 0.7, 0.9)) {
cat(sprintf(" P(w₁ > %.1f) = %.4f\n", t, prob_w1_exceeds(t, a, b)))
}
#> P(w₁ > 0.3) = 0.6634
#> P(w₁ > 0.5) = 0.4868
#> P(w₁ > 0.7) = 0.3334
#> P(w₁ > 0.9) = 0.18332.3 Visualization of the Marginal Distribution
# Visualize the marginal w1 distribution
x_grid <- seq(0.01, 0.99, length.out = 200)
a <- 1.6
b <- 1.22
# Compute density manually using the closed-form formula
# p(w1 | a, b) = a * b^a / ((1-w1) * (b - log(1-w1))^(a+1))
density_w1_manual <- function(x, a, b) {
denom <- (1 - x) * (b - log(1 - x))^(a + 1)
a * b^a / denom
}
w1_df <- data.frame(
x = x_grid,
density = density_w1_manual(x_grid, a, b),
cdf = cdf_w1(x_grid, a, b),
survival = prob_w1_exceeds(x_grid, a, b)
)
p1 <- ggplot(w1_df, aes(x = x, y = density)) +
geom_line(color = "#E41A1C", linewidth = 1) +
geom_vline(xintercept = quantile_w1(0.5, a, b), linetype = "dashed",
color = "#377EB8", alpha = 0.7) +
annotate("text", x = quantile_w1(0.5, a, b) + 0.05, y = max(w1_df$density) * 0.9,
label = "Median", color = "#377EB8", hjust = 0) +
labs(x = expression(w[1]), y = "Density",
title = expression("Marginal Density of " * w[1] * " | Gamma(1.6, 1.22)")) +
theme_minimal()
p2 <- ggplot(w1_df, aes(x = x)) +
geom_line(aes(y = cdf), color = "#377EB8", linewidth = 1) +
geom_line(aes(y = survival), color = "#E41A1C", linewidth = 1, linetype = "dashed") +
geom_hline(yintercept = 0.5, linetype = "dotted", color = "gray50") +
annotate("text", x = 0.9, y = 0.85, label = "CDF", color = "#377EB8") +
annotate("text", x = 0.9, y = 0.35, label = "P(w1 > x)", color = "#E41A1C") +
labs(x = expression(w[1]), y = "Probability",
title = "CDF and Survival Function") +
theme_minimal()
gridExtra::grid.arrange(p1, p2, ncol = 2)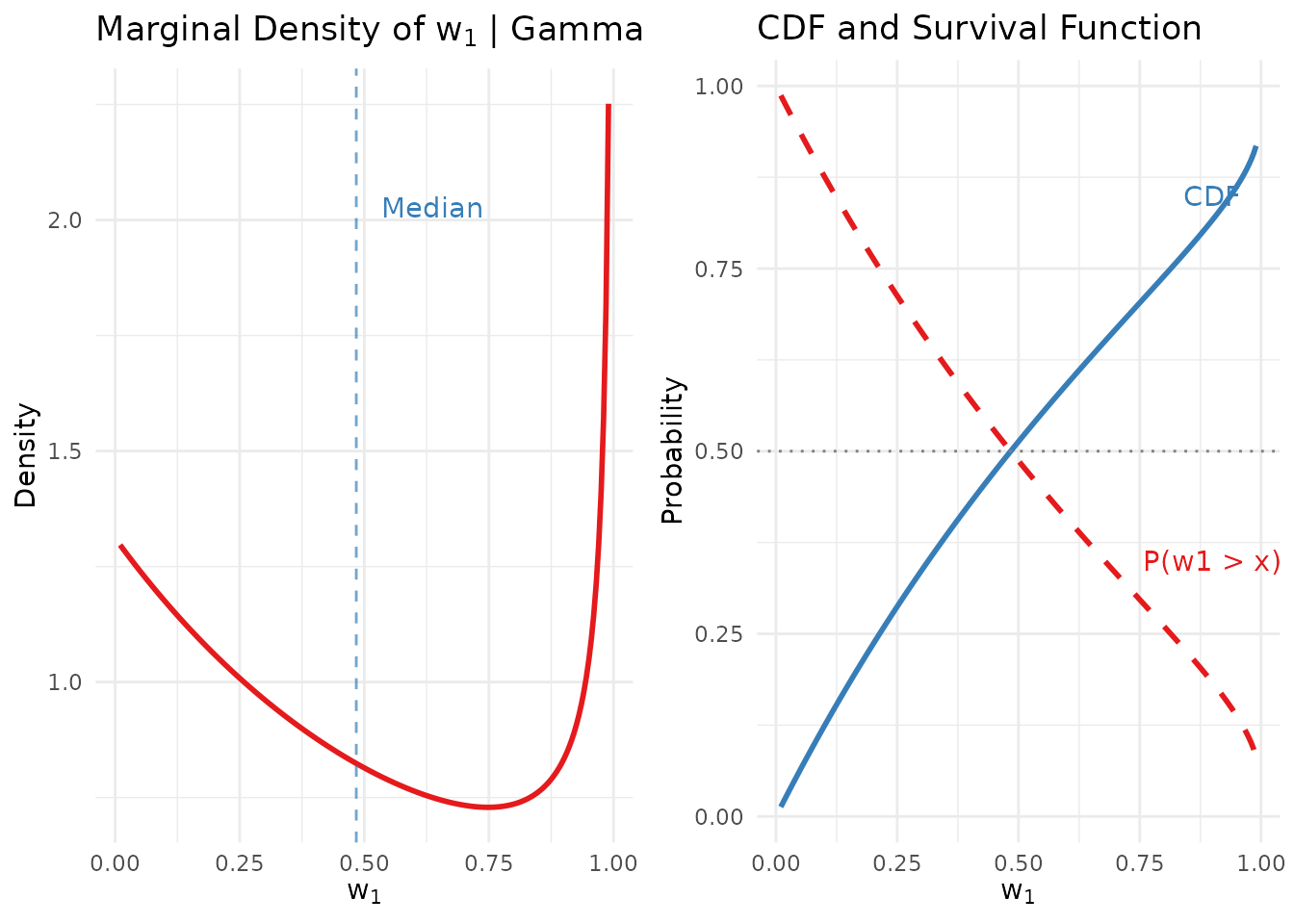
Marginal density and CDF of w₁ under the Gamma(1.6, 1.22) hyperprior.
3. The Co-Clustering Probability
3.1 Definition and Interpretation
The co-clustering probability provides an alternative, often more intuitive, anchor for weight-based elicitation.
Definition 1 (Co-clustering probability / Simpson index).
Interpretation. Conditional on the random measure , if are i.i.d. cluster labels with , then:
Thus is the probability that two randomly chosen units belong to the same latent cluster. This quantity translates directly to applied elicitation:
“Before seeing data, what is the probability that two randomly selected sites from your study belong to the same effect group?”
3.2 Conditional Moments of
Proposition 2 (Conditional expectation of ; Kingman, 1975; Pitman, 1996).
Proof. Using the stick-breaking construction:
For :
Summing the geometric series:
Corollary 1 (Key identity). The conditional expectations of and are equal:
This identity is not coincidental—both quantities measure the “concentration” of the random measure .
Proposition 3 (Conditional variance of ).
Proof. Using the distributional recursion where and is an independent copy of . Squaring and taking expectations yields the result. See Appendix A of the associated research notes for the full derivation.
alpha_grid <- seq(0.1, 20, length.out = 200)
rho_cond_df <- data.frame(
alpha = alpha_grid,
mean = mean_rho_given_alpha(alpha_grid),
var = var_rho_given_alpha(alpha_grid),
sd = sqrt(var_rho_given_alpha(alpha_grid))
)
p1 <- ggplot(rho_cond_df, aes(x = alpha, y = mean)) +
geom_line(color = "#E41A1C", linewidth = 1) +
geom_ribbon(aes(ymin = pmax(0, mean - 2*sd), ymax = pmin(1, mean + 2*sd)),
alpha = 0.2, fill = "#E41A1C") +
labs(x = expression(alpha), y = expression(rho),
title = expression(E*"["*rho*" | "*alpha*"]" * " ± 2 SD")) +
coord_cartesian(ylim = c(0, 1)) +
theme_minimal()
p2 <- ggplot(rho_cond_df, aes(x = alpha, y = var)) +
geom_line(color = "#377EB8", linewidth = 1) +
labs(x = expression(alpha), y = "Variance",
title = expression("Var("*rho*" | "*alpha*")")) +
theme_minimal()
gridExtra::grid.arrange(p1, p2, ncol = 2)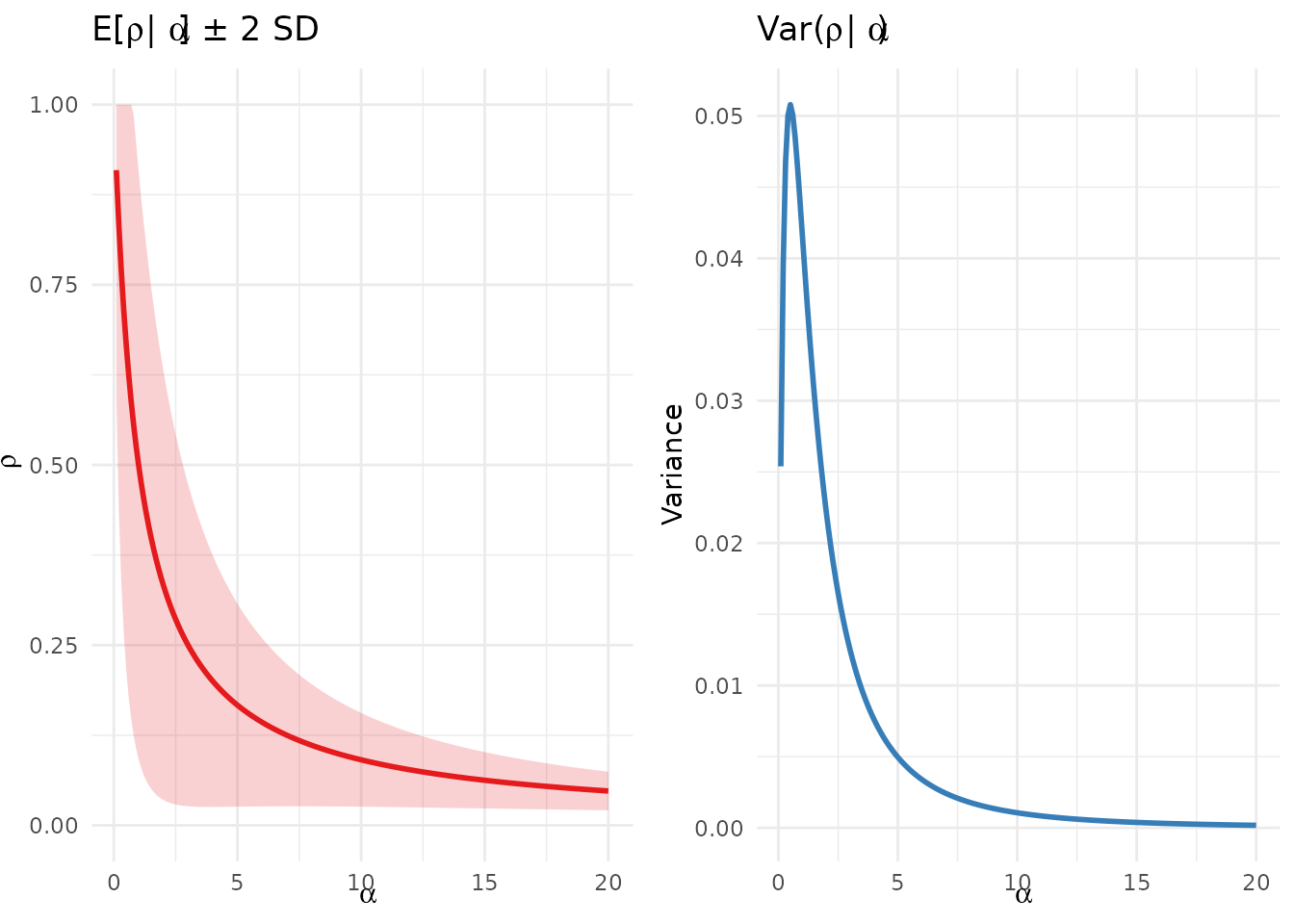
Conditional mean and variance of ρ as functions of α.
4. The Functional: Closed-Form Moments
4.1 Definition and Analytical Expression
Computing unconditional moments of and under a Gamma hyperprior requires evaluating expectations of the form .
Definition 2 (The functional).
Lemma 1 (Closed-form expression for ; this work). where is the upper incomplete gamma function.
Proof. We have:
Using the integral identity: and multiplying by , we obtain the result.
Remark 1 (Numerical evaluation). For
,
the upper incomplete gamma function
is defined via analytic continuation. Standard numerical libraries
handle this automatically (e.g., pgamma in R with
appropriate transformations, or scipy.special.gammaincc in
Python). For practical purposes in the DPprior package, we use
Gauss-Laguerre quadrature which avoids these analytic continuation
subtleties.
4.2 Key Identities
Theorem 2 (Unconditional moments via ; this work).
Let . Then:
Mean of :
Mean of (co-clustering probability):
Corollary 2 (Moment identity). The unconditional means are equal:
This identity has important implications for elicitation: if a practitioner specifies only a target for the mean of or , the two anchors are informationally equivalent. To make add genuine extra constraint, one must elicit uncertainty (e.g., an interval) and match the variance.
# Verify the key identity: E[w1] = E[rho]
test_cases <- list(
c(a = 1.0, b = 1.0),
c(a = 1.6, b = 1.22),
c(a = 2.0, b = 0.5),
c(a = 0.5, b = 2.0)
)
cat("Verification: E[w₁] = E[ρ] identity\n")
#> Verification: E[w₁] = E[ρ] identity
cat(sprintf("%10s %10s %12s %12s %12s\n", "a", "b", "E[w₁]", "E[ρ]", "Difference"))
#> a b E[w₁] E[ρ] Difference
cat(strrep("-", 60), "\n")
#> ------------------------------------------------------------
for (params in test_cases) {
a <- params["a"]
b <- params["b"]
E_w1 <- mean_w1(a, b)
E_rho <- mean_rho(a, b)
diff <- abs(E_w1 - E_rho)
cat(sprintf("%10.2f %10.2f %12.6f %12.6f %12.2e\n", a, b, E_w1, E_rho, diff))
}
#> 1.00 1.00 0.596347 0.596347 0.00e+00
#> 1.60 1.22 0.508368 0.508368 0.00e+00
#> 2.00 0.50 0.269272 0.269272 0.00e+00
#> 0.50 2.00 0.842738 0.842738 0.00e+004.3 Second Moments and Variance of
Proposition 4 (Unconditional second moment of ; this work).
Using the partial fraction decomposition:
we obtain:
Hence the unconditional variance:
Key design implication. The distribution of is not closed-form, but mean and variance are closed-form (“moment-closed”). This is sufficient for moment-based calibration and diagnostics.
# Compute unconditional rho moments for the Lee et al. prior
a <- 1.6
b <- 1.22
cat("Co-clustering probability under Gamma(1.6, 1.22) hyperprior:\n")
#> Co-clustering probability under Gamma(1.6, 1.22) hyperprior:
cat(sprintf(" E[ρ] = %.4f\n", mean_rho(a, b)))
#> E[ρ] = 0.5084
cat(sprintf(" Var(ρ) = %.4f\n", var_rho(a, b)))
#> Var(ρ) = 0.0710
cat(sprintf(" SD(ρ) = %.4f\n", sqrt(var_rho(a, b))))
#> SD(ρ) = 0.2664
cat(sprintf(" CV(ρ) = %.4f\n", cv_rho(a, b)))
#> CV(ρ) = 0.52405. The Dual-Anchor Optimization Framework
5.1 Motivation: Why K-Only Calibration is Insufficient
As demonstrated by Vicentini and Jermyn (2025, Section 5), priors calibrated to match a target on the number of clusters can induce materially informative priors on the stick-breaking weights. They observe that “-diffuse, DORO, quasi-degenerate and Jeffreys’ priors… are markedly different from the behaviour of SSI” (p. 18).
The mechanism is structural: and the weight distribution are controlled by different functionals of . For fixed : while:
Matching the location of pins down around , but this implied can correspond to high dominance risk in .
# Demonstrate the unintended prior problem
J <- 50
mu_K_target <- 5
# Fit using K-only calibration
fit_K <- DPprior_fit(J = J, mu_K = mu_K_target, confidence = "medium")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 49.7% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
# What does this imply for weights?
cat("K-only calibration (J=50, μ_K=5):\n")
#> K-only calibration (J=50, μ_K=5):
cat(sprintf(" Gamma hyperprior: a = %.3f, b = %.3f\n", fit_K$a, fit_K$b))
#> Gamma hyperprior: a = 1.408, b = 1.077
cat(sprintf(" Achieved E[K] = %.2f\n",
exact_K_moments(J, fit_K$a, fit_K$b)$mean))
#> Achieved E[K] = 5.00
cat("\nImplied weight behavior:\n")
#>
#> Implied weight behavior:
cat(sprintf(" E[w₁] = %.3f\n", mean_w1(fit_K$a, fit_K$b)))
#> E[w₁] = 0.518
cat(sprintf(" P(w₁ > 0.5) = %.3f\n", prob_w1_exceeds(0.5, fit_K$a, fit_K$b)))
#> P(w₁ > 0.5) = 0.497
cat(sprintf(" P(w₁ > 0.9) = %.3f\n", prob_w1_exceeds(0.9, fit_K$a, fit_K$b)))
#> P(w₁ > 0.9) = 0.200
# Visualize the implied w1 distribution
x_grid <- seq(0.01, 0.99, length.out = 200)
implied_df <- data.frame(
x = x_grid,
density = density_w1_manual(x_grid, fit_K$a, fit_K$b),
type = "K-only calibration"
)
ggplot(implied_df, aes(x = x, y = density)) +
geom_line(color = "#377EB8", linewidth = 1.2) +
geom_vline(xintercept = 0.5, linetype = "dashed", color = "#E41A1C", alpha = 0.7) +
annotate("text", x = 0.52, y = max(implied_df$density) * 0.8,
label = "50% threshold", color = "#E41A1C", hjust = 0) +
labs(
x = expression(w[1]),
y = "Density",
title = expression("Implied Distribution of " * w[1] * " Under K-Only Calibration"),
subtitle = sprintf("J = %d, target E[K] = %d; P(w₁ > 0.5) = %.2f",
J, mu_K_target, prob_w1_exceeds(0.5, fit_K$a, fit_K$b))
) +
theme_minimal()![The K-calibrated prior (blue) achieves the target E[K] but implies higher dominance risk than practitioners might expect.](theory-weights_files/figure-html/unintended-prior-demo-1.png)
The K-calibrated prior (blue) achieves the target E[K] but implies higher dominance risk than practitioners might expect.
5.2 The Dual-Anchor Objective
Definition 3 (Dual-anchor optimization; this work).
Given elicited targets (or summary statistics thereof) and where , the dual-anchor hyperparameters are: where: and controls the trade-off between the two anchors.
Boundary cases:
| Behavior | |
|---|---|
| K-only calibration (RN-01 through RN-05) | |
| Weight-only calibration (SSI-style) | |
| Compromise between both anchors |
5.3 Implementation Variants
The dual-anchor objective can be implemented using different loss functions:
Moment-based loss (recommended for stability): where are tuning weights.
Quantile-based loss for : For , one can directly target specific quantiles using the closed-form : where are elicited quantile pairs.
Probability constraint for : Target a specific dominance risk:
# Demonstrate dual-anchor calibration
fit_dual <- DPprior_dual(
fit = fit_K,
w1_target = list(prob = list(threshold = 0.5, value = 0.25)),
lambda = 0.7,
loss_type = "adaptive",
verbose = FALSE
)
cat("Dual-anchor calibration (target: P(w₁ > 0.5) = 0.25):\n")
#> Dual-anchor calibration (target: P(w₁ > 0.5) = 0.25):
cat(sprintf(" Gamma hyperprior: a = %.3f, b = %.3f\n", fit_dual$a, fit_dual$b))
#> Gamma hyperprior: a = 3.078, b = 1.669
cat(sprintf(" Achieved E[K] = %.2f (target: %.1f)\n",
exact_K_moments(J, fit_dual$a, fit_dual$b)$mean, mu_K_target))
#> Achieved E[K] = 6.43 (target: 5.0)
cat(sprintf(" P(w₁ > 0.5) = %.3f (target: 0.25)\n",
prob_w1_exceeds(0.5, fit_dual$a, fit_dual$b)))
#> P(w₁ > 0.5) = 0.343 (target: 0.25)5.4 The Trade-off Curve
# Compute trade-off curve
lambda_grid <- c(1.0, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3)
w1_target <- list(prob = list(threshold = 0.5, value = 0.2))
tradeoff_results <- lapply(lambda_grid, function(lam) {
if (lam == 1.0) {
fit <- fit_K
fit$dual_anchor <- list(lambda = 1.0)
} else {
fit <- DPprior_dual(fit_K, w1_target, lambda = lam,
loss_type = "adaptive", verbose = FALSE)
}
list(
lambda = lam,
a = fit$a,
b = fit$b,
E_K = exact_K_moments(J, fit$a, fit$b)$mean,
P_w1_gt_50 = prob_w1_exceeds(0.5, fit$a, fit$b),
E_w1 = mean_w1(fit$a, fit$b)
)
})
tradeoff_df <- data.frame(
lambda = sapply(tradeoff_results, `[[`, "lambda"),
E_K = sapply(tradeoff_results, `[[`, "E_K"),
P_w1 = sapply(tradeoff_results, `[[`, "P_w1_gt_50"),
E_w1 = sapply(tradeoff_results, `[[`, "E_w1")
)
ggplot(tradeoff_df, aes(x = E_K, y = P_w1)) +
geom_path(color = "gray50", linewidth = 0.8) +
geom_point(aes(color = factor(lambda)), size = 4) +
geom_hline(yintercept = 0.2, linetype = "dashed", color = "#E41A1C", alpha = 0.7) +
geom_vline(xintercept = 5, linetype = "dashed", color = "#377EB8", alpha = 0.7) +
annotate("text", x = 4.5, y = 0.22, label = "w1 target",
hjust = 1, color = "#E41A1C") +
annotate("text", x = 5.1, y = 0.45, label = "K target", hjust = 0, color = "#377EB8") +
scale_color_viridis_d(option = "viridis", begin = 0.2, end = 0.8) +
labs(
x = expression(E*"["*K[J]*"]"),
y = expression(P(w[1] > 0.5)),
title = "Dual-Anchor Trade-off Curve",
subtitle = "Varying λ from 1.0 (K-only) to 0.3 (weight-priority)",
color = "λ"
) +
theme_minimal() +
theme(legend.position = "right")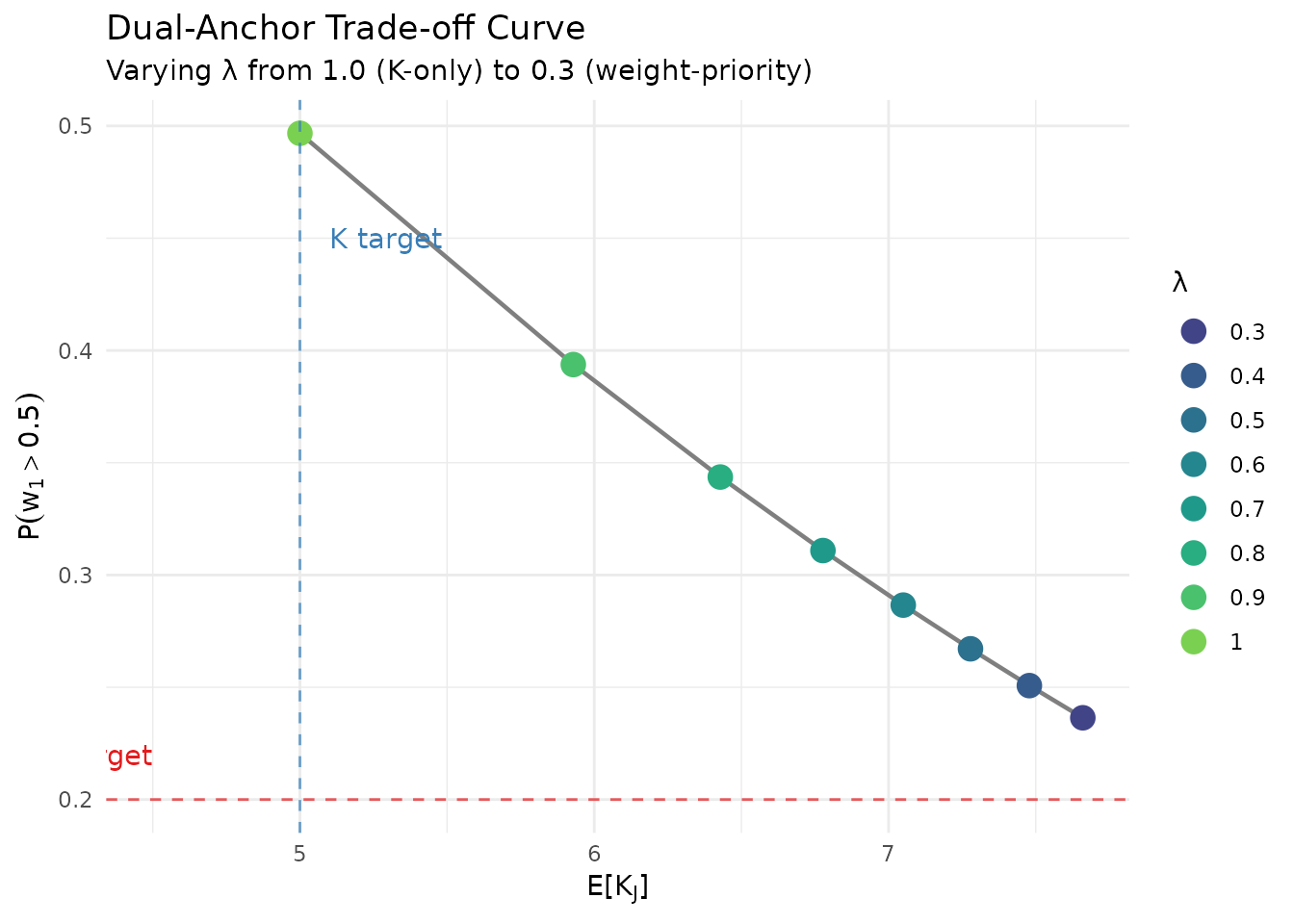
Trade-off between K-fit and weight control as λ varies from 1 (K-only) to 0.3 (weight-priority).
6. Relationship Between and
6.1 When Are They Equivalent?
As established above, . This extends to the unconditional case: .
Consequence: If the practitioner elicits only a mean constraint (e.g., “co-clustering probability should be about 0.3”), using or as the second anchor yields identical calibration constraints.
6.2 When Are They Different?
The variances differ:
alpha_grid <- seq(0.1, 20, length.out = 200)
var_comparison <- data.frame(
alpha = rep(alpha_grid, 2),
variance = c(
alpha_grid / ((1 + alpha_grid)^2 * (2 + alpha_grid)), # Var(w1|alpha)
var_rho_given_alpha(alpha_grid) # Var(rho|alpha)
),
quantity = rep(c("Var(w₁ | α)", "Var(ρ | α)"), each = length(alpha_grid))
)
ggplot(var_comparison, aes(x = alpha, y = variance, color = quantity)) +
geom_line(linewidth = 1) +
scale_color_manual(values = c("#E41A1C", "#377EB8")) +
labs(
x = expression(alpha),
y = "Conditional Variance",
title = expression("Conditional Variance Comparison: " * w[1] * " vs " * rho),
color = NULL
) +
theme_minimal() +
theme(legend.position = "bottom")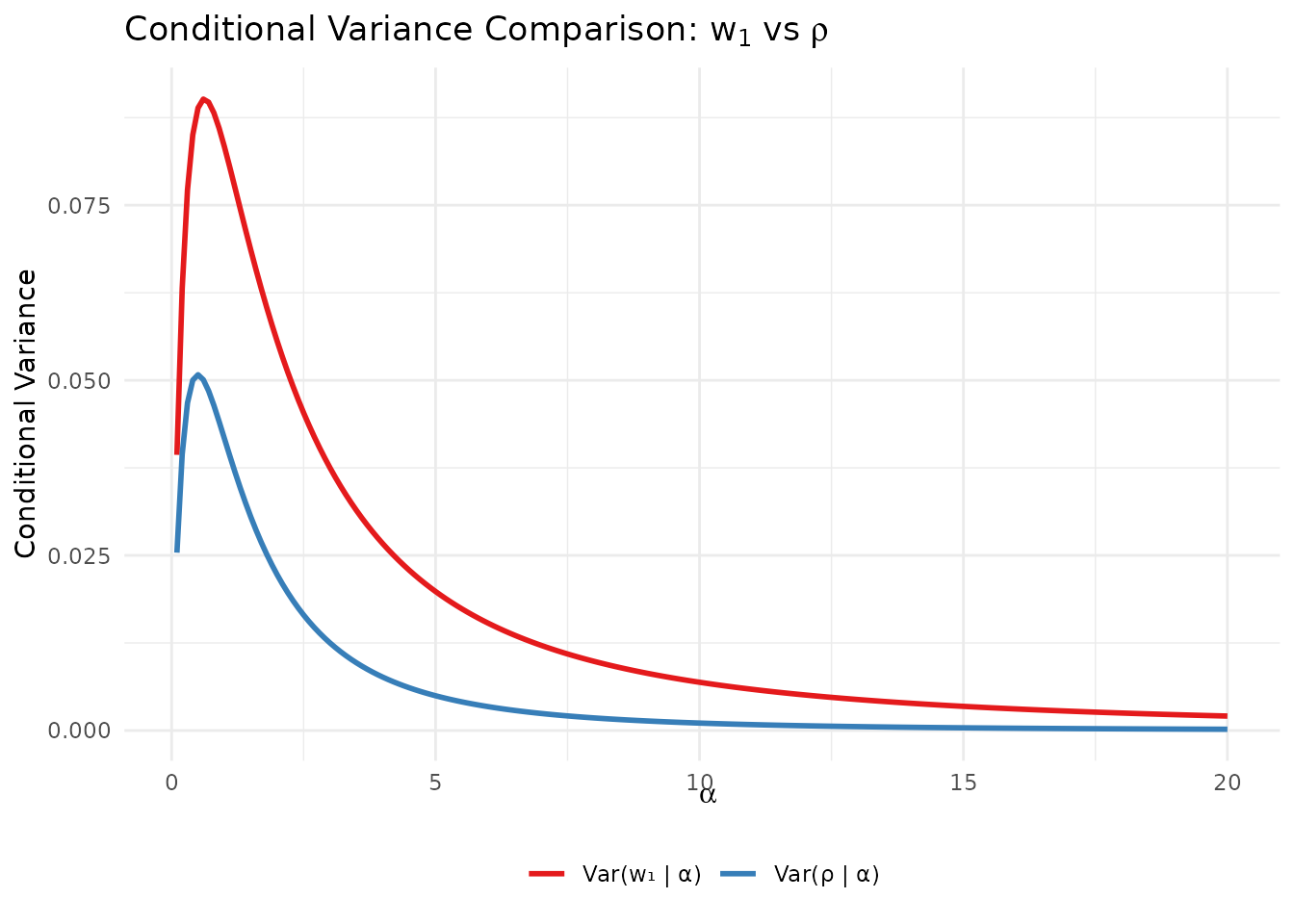
Comparison of conditional variances: Var(w₁|α) vs Var(ρ|α).
6.3 Practical Guidance: Choosing Between and
| Criterion | ||
|---|---|---|
| Distribution tractability | Fully closed-form (CDF, quantiles) | Moment-closed only |
| Elicitation intuitiveness | Moderate | High (“probability two share a cluster”) |
| Tail constraints | Direct via | Requires simulation |
| Variance matching | Possible but complex | Straightforward |
Recommendation: Use for quantile/probability constraints; use when the co-clustering interpretation resonates with domain experts.
7. Computational Details
7.1 Key Functions in the DPprior Package
The package provides efficient implementations for all weight-related computations:
| Function | Description |
|---|---|
mean_w1(a, b) |
via Gauss-Laguerre quadrature |
var_w1(a, b) |
via quadrature |
cdf_w1(x, a, b) |
Closed-form CDF |
quantile_w1(u, a, b) |
Closed-form quantile function |
prob_w1_exceeds(t, a, b) |
Closed-form |
mean_rho(a, b) |
via quadrature |
var_rho(a, b) |
via quadrature |
mean_rho_given_alpha(alpha) |
Conditional mean |
var_rho_given_alpha(alpha) |
Conditional variance |
DPprior_dual() |
Dual-anchor calibration |
7.2 Numerical Verification
# Verify implementations against Monte Carlo
a <- 1.6
b <- 1.22
n_mc <- 100000
set.seed(42)
alpha_samples <- rgamma(n_mc, shape = a, rate = b)
w1_samples <- rbeta(n_mc, 1, alpha_samples)
cat("Verification against Monte Carlo (n =", format(n_mc, big.mark = ","), "):\n\n")
#> Verification against Monte Carlo (n = 1e+05 ):
cat("w₁ moments:\n")
#> w₁ moments:
cat(sprintf(" E[w₁]: Analytic = %.4f, MC = %.4f\n",
mean_w1(a, b), mean(w1_samples)))
#> E[w₁]: Analytic = 0.5084, MC = 0.5088
cat(sprintf(" Var(w₁): Analytic = %.4f, MC = %.4f\n",
var_w1(a, b), var(w1_samples)))
#> Var(w₁): Analytic = 0.1052, MC = 0.1054
cat("\nw₁ tail probabilities:\n")
#>
#> w₁ tail probabilities:
for (t in c(0.3, 0.5, 0.9)) {
mc_prob <- mean(w1_samples > t)
analytic_prob <- prob_w1_exceeds(t, a, b)
cat(sprintf(" P(w₁ > %.1f): Analytic = %.4f, MC = %.4f\n",
t, analytic_prob, mc_prob))
}
#> P(w₁ > 0.3): Analytic = 0.6634, MC = 0.6627
#> P(w₁ > 0.5): Analytic = 0.4868, MC = 0.4876
#> P(w₁ > 0.9): Analytic = 0.1833, MC = 0.1858
cat("\nρ moments (via quadrature vs MC simulation):\n")
#>
#> ρ moments (via quadrature vs MC simulation):
rho_samples <- sapply(alpha_samples, function(alph) {
v <- rbeta(100, 1, alph)
w <- cumprod(c(1, 1 - v[-length(v)])) * v
sum(w^2)
})
cat(sprintf(" E[ρ]: Analytic = %.4f, MC = %.4f\n",
mean_rho(a, b), mean(rho_samples)))
#> E[ρ]: Analytic = 0.5084, MC = 0.50818. Summary
This vignette has provided a rigorous treatment of the weight distribution theory underlying the DPprior package:
Stick-breaking weights follow the GEM() distribution in size-biased (not decreasing) order.
, and under , the marginal distribution of has fully closed-form CDF, quantiles, and tail probabilities.
The co-clustering probability has the same conditional mean as : .
The functional enables closed-form computation of , which underlies the moment formulas for both and .
K-only calibration can induce unintended weight behavior. The dual-anchor framework provides explicit control over both cluster counts and weight concentration via a trade-off parameter .
References
Antoniak, C. E. (1974). Mixtures of Dirichlet processes with applications to Bayesian nonparametric problems. The Annals of Statistics, 2(6), 1152-1174.
Arratia, R., Barbour, A. D., & Tavaré, S. (2003). Logarithmic Combinatorial Structures: A Probabilistic Approach. European Mathematical Society.
Connor, R. J., & Mosimann, J. E. (1969). Concepts of independence for proportions with a generalization of the Dirichlet distribution. Journal of the American Statistical Association, 64(325), 194-206.
Dorazio, R. M. (2009). On selecting a prior for the precision parameter of Dirichlet process mixture models. Journal of Statistical Planning and Inference, 139(10), 3384-3390.
Escobar, M. D., & West, M. (1995). Bayesian density estimation and inference using mixtures. Journal of the American Statistical Association, 90(430), 577-588.
Kingman, J. F. C. (1975). Random discrete distributions. Journal of the Royal Statistical Society: Series B, 37(1), 1-22.
Lee, J., Che, J., Rabe-Hesketh, S., Feller, A., & Miratrix, L. (2025). Improving the estimation of site-specific effects and their distribution in multisite trials. Journal of Educational and Behavioral Statistics, 50(5), 731-764.
Murugiah, S., & Sweeting, T. J. (2012). Selecting the precision parameter prior in Dirichlet process mixture models. Journal of Statistical Planning and Inference, 142(7), 1947-1959.
Pitman, J. (1996). Random discrete distributions invariant under size-biased permutation. Advances in Applied Probability, 28(2), 525-539.
Sethuraman, J. (1994). A constructive definition of Dirichlet priors. Statistica Sinica, 4(2), 639-650.
Vicentini, S., & Jermyn, I. H. (2025). Prior selection for the precision parameter of Dirichlet process mixture models. arXiv:2502.00864.
Zito, A., Rigon, T., & Dunson, D. B. (2024). Bayesian nonparametric modeling of latent partitions via Stirling-gamma priors. Bayesian Analysis. https://doi.org/10.1214/24-BA1463
Appendix: Key Formulas Reference
A.3 The Functional
For questions about this vignette or the DPprior package, please visit the GitHub repository.