Case Studies: Multisite Trials and Meta-Analysis
JoonHo Lee
2026-01-01
Source:vignettes/case-studies.Rmd
case-studies.RmdOverview
This vignette presents three case studies demonstrating how to apply the DPprior package in real-world research contexts. Each case study walks through the complete workflow—from substantive considerations to final prior specification—using published research as a foundation.
The three case studies span different domains and data structures:
| Case Study | Domain | J (Sites/Studies) | Key Characteristic |
|---|---|---|---|
| 1. Conditional Cash Transfer Trial | Education/Policy | 38 | Moderate sites, dual-anchor refinement |
| 2. Brief Alcohol Interventions | Public Health | 117 | Large meta-analysis, multiple heterogeneity sources |
| 3. Alabama Pre-K Value-Added | Education Policy | 500 (of 847) | Large-scale policy evaluation |
By working through these examples, you will learn how to:
- Translate substantive domain knowledge into cluster count expectations
- Choose appropriate calibration methods for different settings
- Apply diagnostics to verify prior behavior
- Conduct sensitivity analysis across specifications
- Report prior elicitation transparently in publications
1. Case Study: Conditional Cash Transfer Multisite Trial
1.1 Research Context
Our first case study draws from the multisite conditional cash transfer (CCT) experiment analyzed in Lee et al. (2025). The Conditional Subsidies for School Attendance program, implemented in Bogotá, Colombia (Barrera-Osorio et al., 2019), conducted randomized experiments across 38 sites in the San Cristobal district.
Study characteristics:
- Sample size: 6,506 participants nested within 38 sites
- Average site size: 171.2 participants
- Coefficient of variation in site sizes: 0.67 (range: 23 to 484)
- Outcomes: Secondary school enrollment and on-time graduation
- Key feature: Large within-site information, limited between-site variation
1.2 The Inferential Goals
The researchers sought to understand the distribution of site-specific treatment effects , with particular interest in:
- Estimating the variance of the prior distribution
- Identifying extreme sites (10th and 90th percentiles)
- Understanding heterogeneity patterns across sites
Given the estimated cross-site effect standard deviation of to (in effect size units) and relatively high within-site precision, this setting exhibits small between-site information and large within-site information—a configuration where prior choice substantially influences posterior inference.
1.3 Initial Prior Elicitation
Step 1: Determining the expected number of clusters
When eliciting prior beliefs about cluster structure, researchers might consider several substantive factors:
- The intervention was implemented uniformly across sites, suggesting limited structural heterogeneity
- However, local context (school culture, neighborhood characteristics) might create differential responses
- Previous multisite education trials typically exhibit 3–8 distinct response patterns
Based on these considerations, a reasonable expectation is approximately clusters among the 38 sites.
# Define the study context
J_cct <- 38
mu_K_cct <- 5
cat("Study context:\n")
#> Study context:
cat(" Number of sites (J):", J_cct, "\n")
#> Number of sites (J): 38
cat(" Expected clusters (μ_K):", mu_K_cct, "\n")
#> Expected clusters (μ_K): 5
cat(" Ratio (μ_K/J):", round(mu_K_cct / J_cct, 3), "\n")
#> Ratio (μ_K/J): 0.132Step 2: Expressing uncertainty
Given the novelty of this specific intervention context and the moderate sample of sites, we should express substantial uncertainty. A “low” confidence level is appropriate:
# Initial fit with low confidence
fit_cct <- DPprior_fit(
J = J_cct,
mu_K = mu_K_cct,
confidence = "low"
)
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 54.5% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
print(fit_cct)
#> DPprior Prior Elicitation Result
#> =============================================
#>
#> Gamma Hyperprior: α ~ Gamma(a = 0.4629, b = 0.2556)
#> E[α] = 1.811, SD[α] = 2.662
#>
#> Target (J = 38):
#> E[K_J] = 5.00
#> Var(K_J) = 20.00
#> (from confidence = 'low')
#>
#> Achieved:
#> E[K_J] = 5.000000, Var(K_J) = 20.000000
#> Residual = 6.09e-09
#>
#> Method: A2-MN (11 iterations)
#>
#> Dominance Risk: HIGH ✘ (P(w₁>0.5) = 54%)1.4 Comprehensive Diagnostics
Before accepting this prior, we need to verify its behavior across
multiple dimensions. The DPprior_diagnostics() function
provides a comprehensive assessment:
diag_cct <- DPprior_diagnostics(fit_cct)
cat("=== Comprehensive Diagnostic Report ===\n\n")
#> === Comprehensive Diagnostic Report ===
cat("1. ALPHA DISTRIBUTION\n")
#> 1. ALPHA DISTRIBUTION
cat(" Mean: ", round(diag_cct$alpha$mean, 3), "\n")
#> Mean: 1.811
cat(" SD: ", round(diag_cct$alpha$sd, 3), "\n")
#> SD: 2.662
cat(" CV: ", round(diag_cct$alpha$cv, 3), "\n")
#> CV: 1.47
cat(" 5th %ile: ", round(diag_cct$alpha$quantiles["q5"], 3), "\n")
#> 5th %ile: 0.005
cat(" 50th %ile:", round(diag_cct$alpha$quantiles["q50"], 3), "\n")
#> 50th %ile: 0.767
cat(" 95th %ile:", round(diag_cct$alpha$quantiles["q95"], 3), "\n")
#> 95th %ile: 7.15
cat("\n2. CLUSTER COUNT (K)\n")
#>
#> 2. CLUSTER COUNT (K)
cat(" E[K]: ", round(diag_cct$K$mean, 3), "\n")
#> E[K]: 5
cat(" Var(K): ", round(diag_cct$K$var, 3), "\n")
#> Var(K): 20
cat(" SD(K): ", round(sqrt(diag_cct$K$var), 3), "\n")
#> SD(K): 4.472
cat(" Mode(K): ", diag_cct$K$mode, "\n")
#> Mode(K): 1
cat("\n3. WEIGHT BEHAVIOR\n")
#>
#> 3. WEIGHT BEHAVIOR
cat(" E[w₁]: ", round(diag_cct$weights$mean, 3), "\n")
#> E[w₁]: 0.572
cat(" Median(w₁): ", round(diag_cct$weights$median, 3), "\n")
#> Median(w₁): 0.588
cat(" P(w₁ > 0.3): ", round(diag_cct$weights$prob_exceeds["prob_gt_0.3"], 3), "\n")
#> P(w₁ > 0.3): NA
cat(" P(w₁ > 0.5): ", round(diag_cct$weights$prob_exceeds["prob_gt_0.5"], 3), "\n")
#> P(w₁ > 0.5): 0.545
cat(" P(w₁ > 0.9): ", round(diag_cct$weights$prob_exceeds["prob_gt_0.9"], 3), "\n")
#> P(w₁ > 0.9): 0.344
cat(" Dominance: ", toupper(diag_cct$weights$dominance_risk), "\n")
#> Dominance: HIGH
cat("\n4. CO-CLUSTERING PROBABILITY (ρ)\n")
#>
#> 4. CO-CLUSTERING PROBABILITY (ρ)
cat(" E[ρ]: ", round(diag_cct$coclustering$mean, 3), "\n")
#> E[ρ]: 0.572
cat(" Interpretation: Two random sites have a",
round(100 * diag_cct$coclustering$mean), "% chance of sharing a cluster\n")
#> Interpretation: Two random sites have a 57 % chance of sharing a cluster1.5 Visualizing the Initial Prior
The DPprior package provides several visualization functions to help understand prior behavior. Let’s examine each component:
Alpha distribution:
plot_alpha_prior(fit_cct)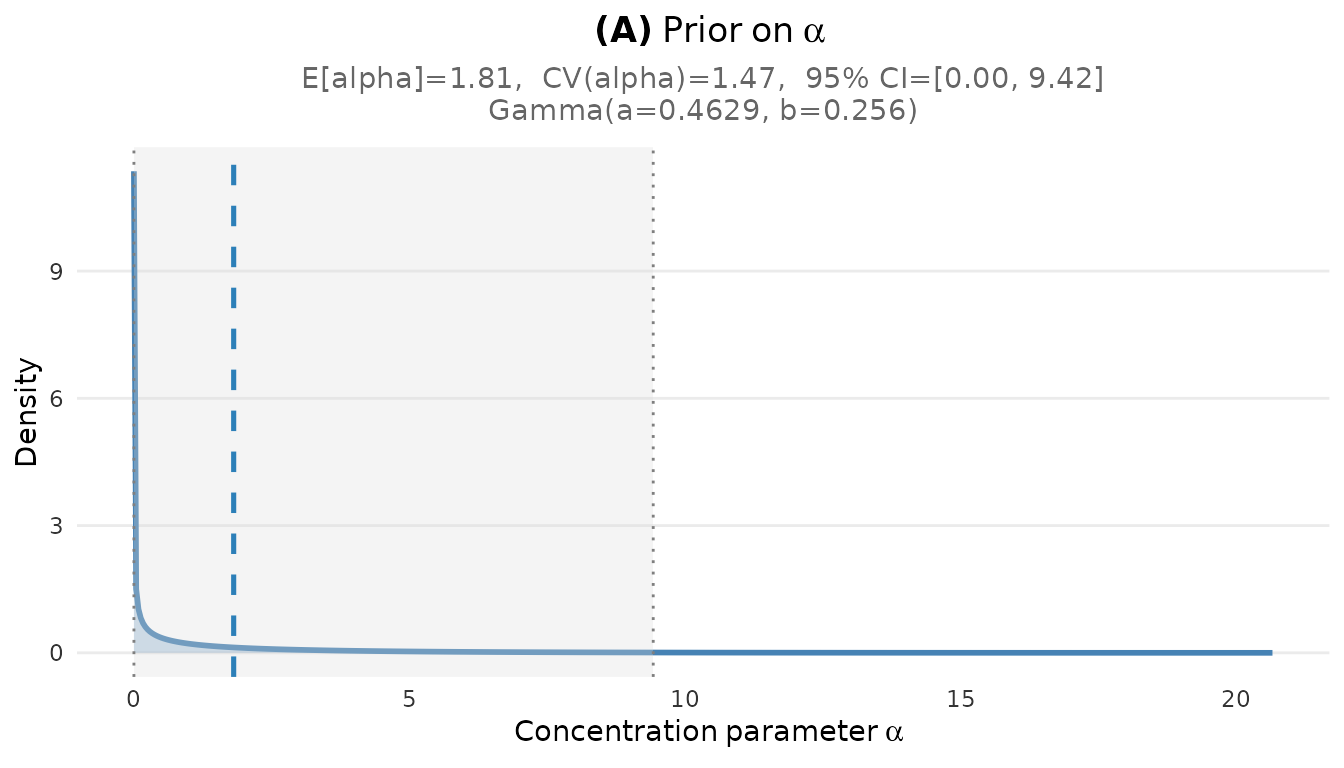
Prior distribution on the concentration parameter α for the CCT study.
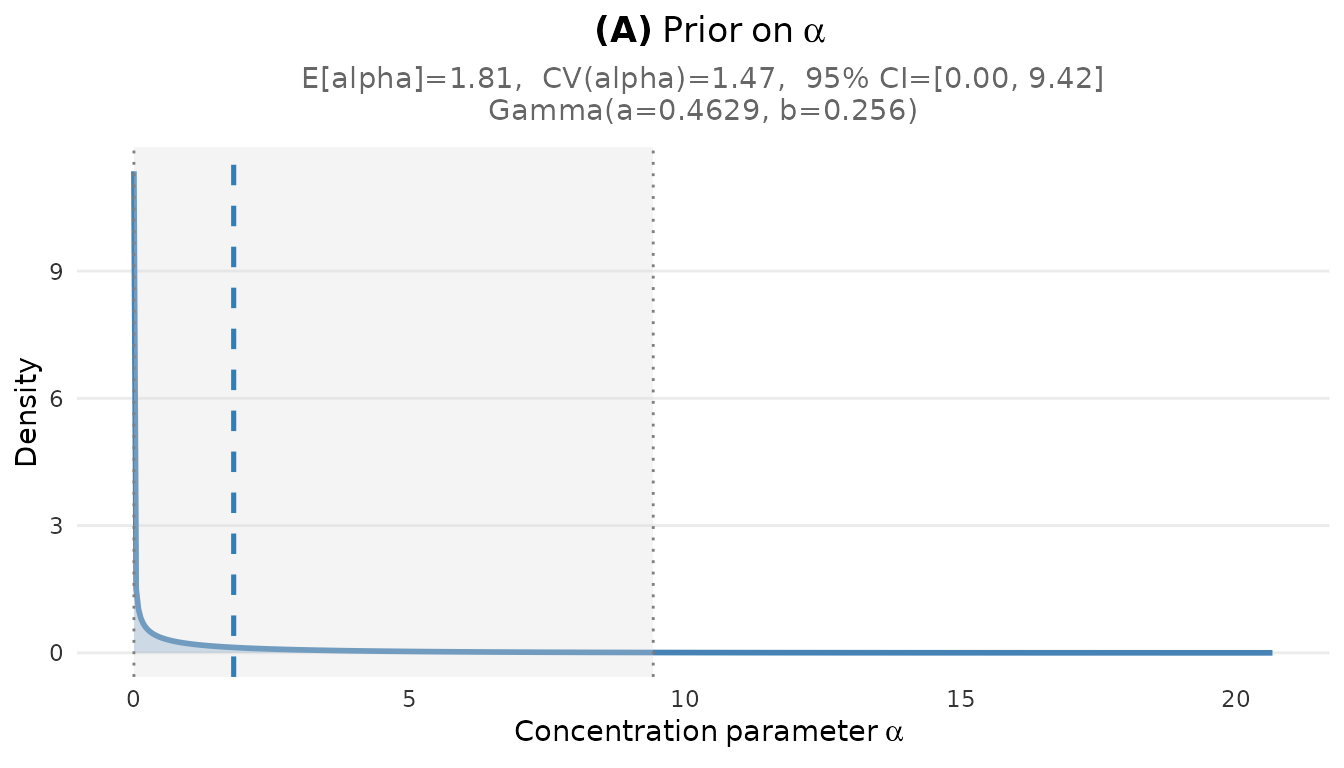
Prior distribution on the concentration parameter α for the CCT study.
Cluster count distribution:
plot_K_prior(fit_cct)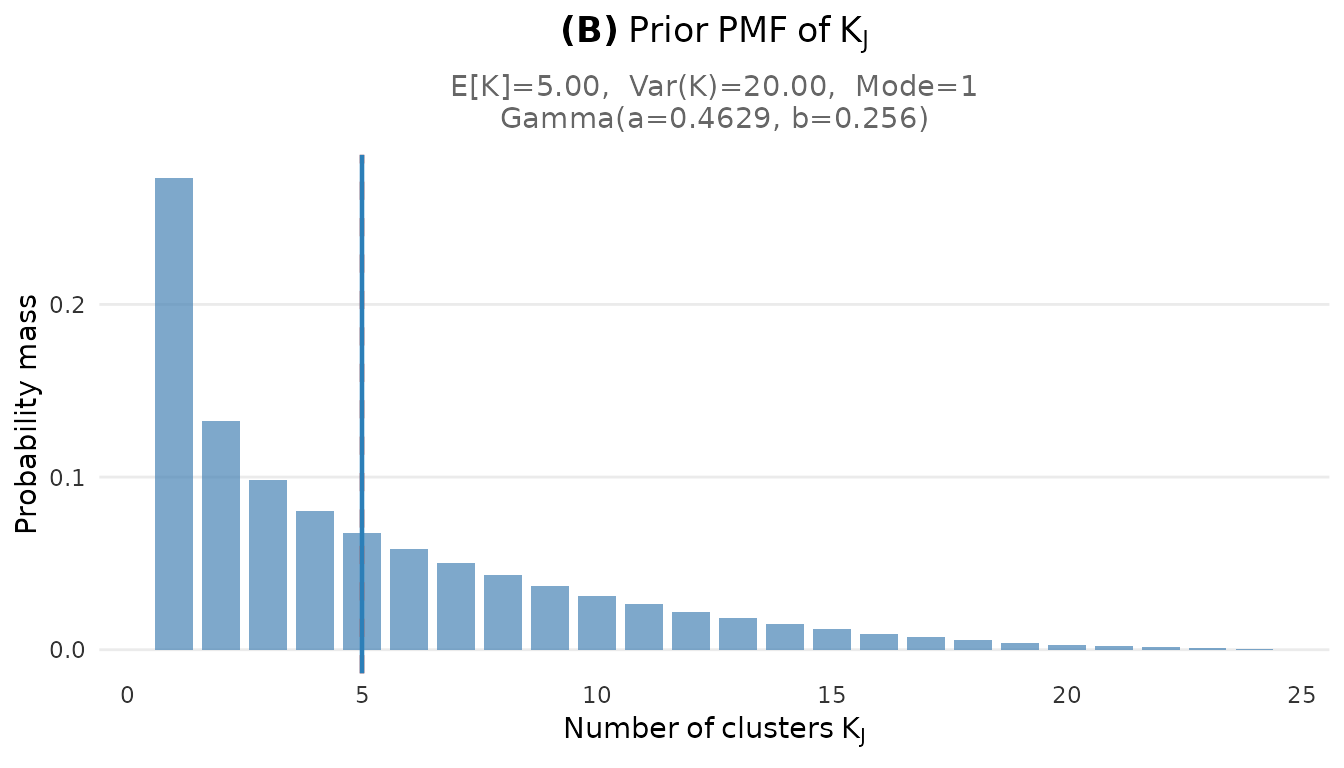
Prior PMF of the number of clusters K for the CCT study (J = 38).
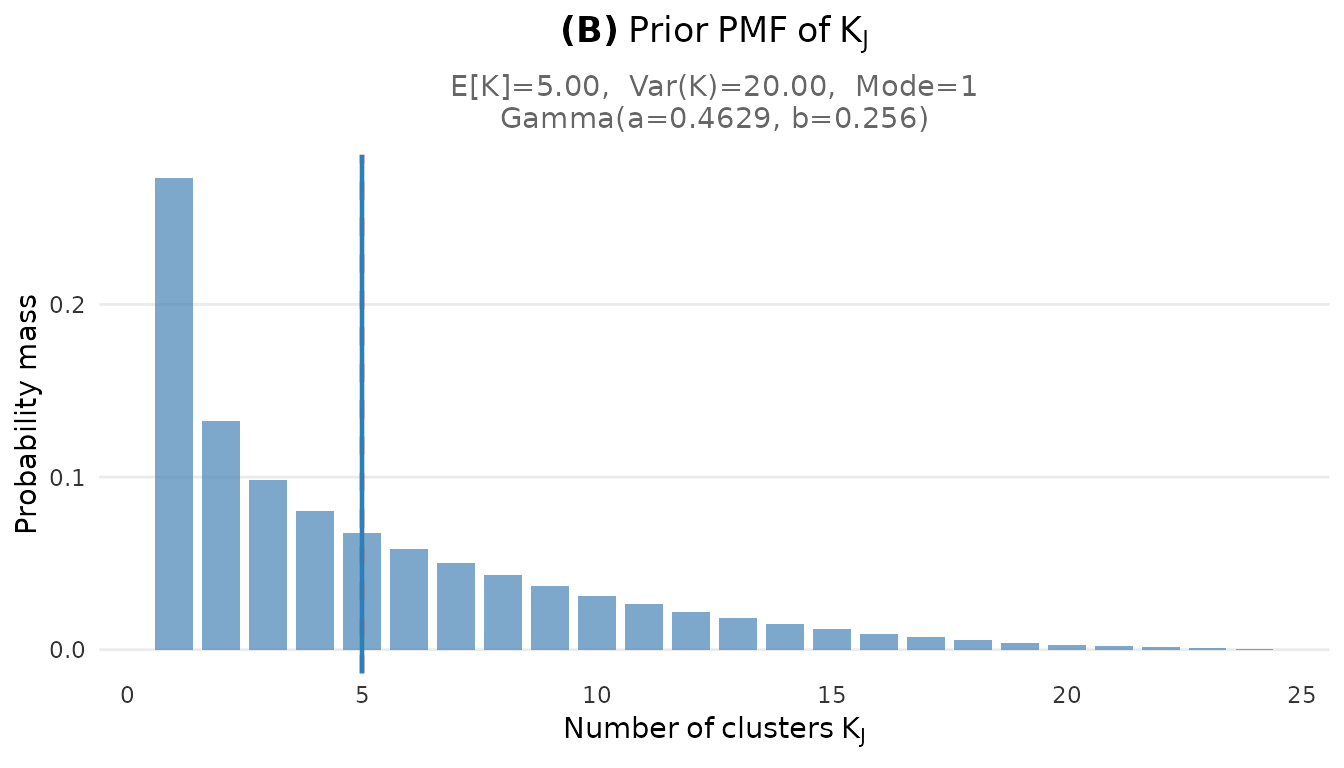
Prior PMF of the number of clusters K for the CCT study (J = 38).
Largest cluster weight distribution:
plot_w1_prior(fit_cct)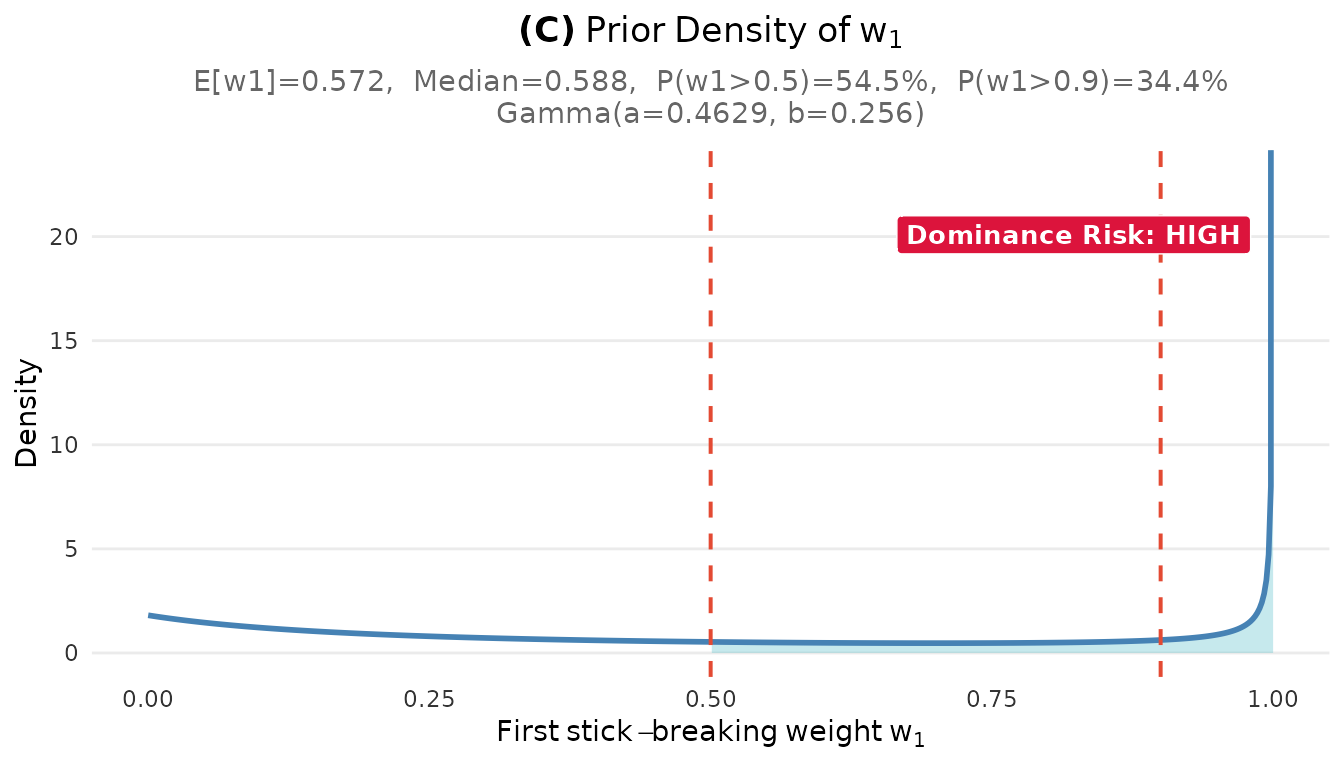
Prior distribution on the largest cluster weight w₁. The dashed line at 0.5 indicates the dominance threshold.
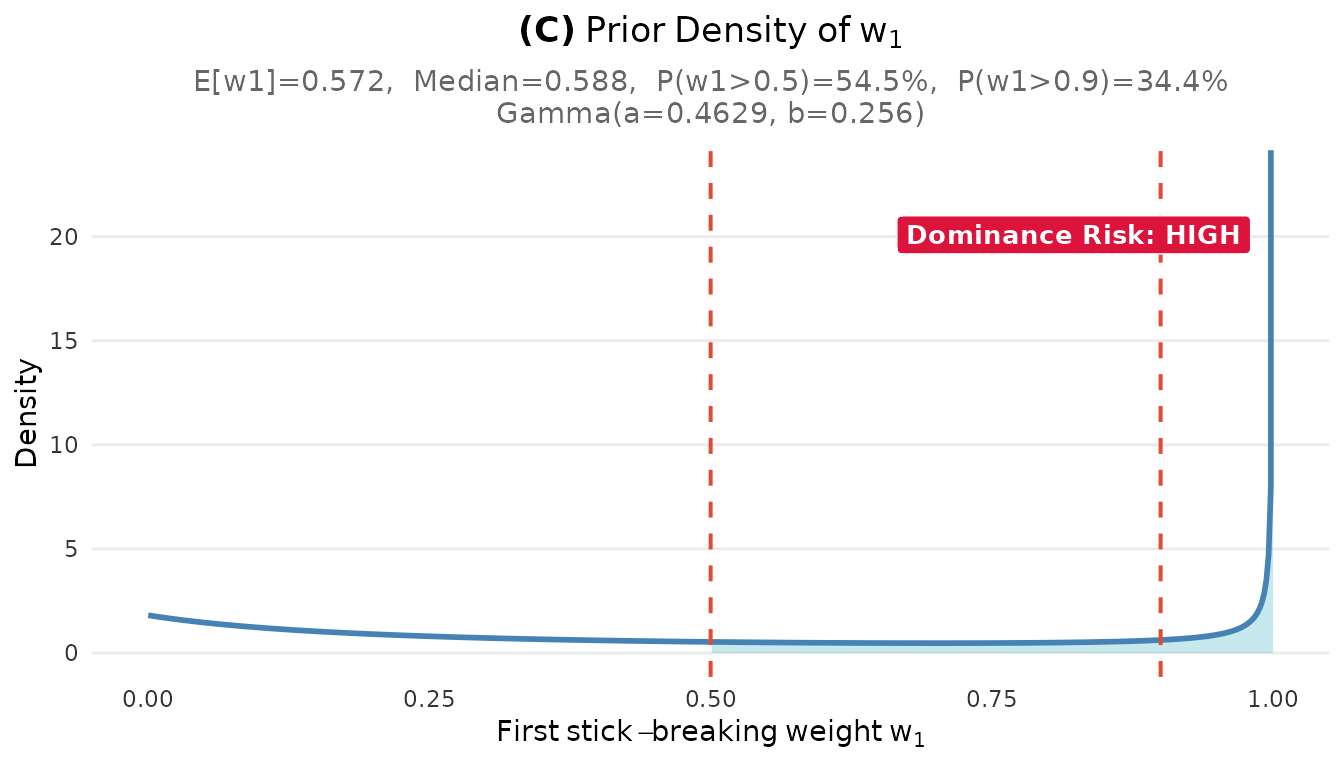
Prior distribution on the largest cluster weight w₁. The dashed line at 0.5 indicates the dominance threshold.
Complete dashboard:
plot(fit_cct)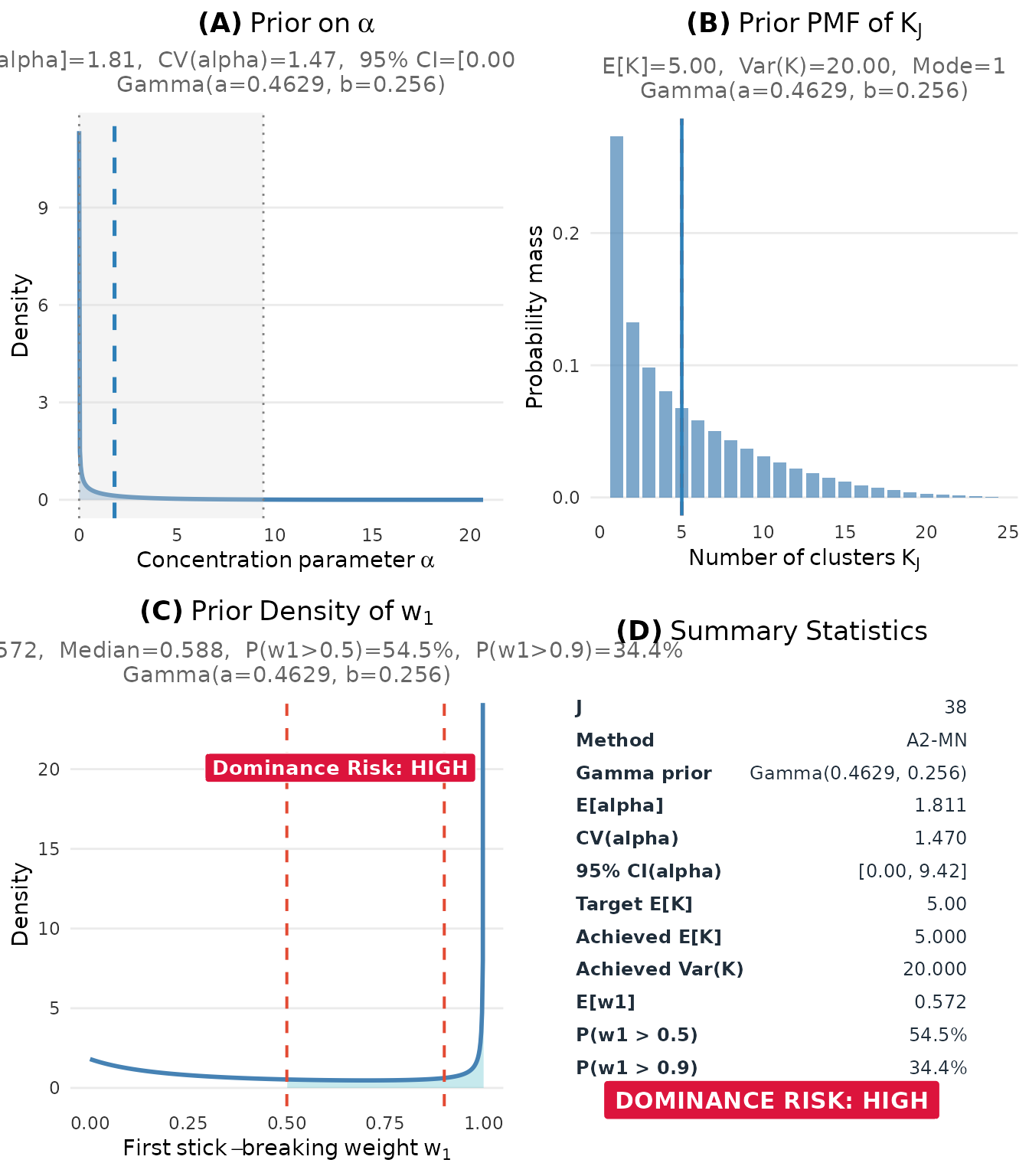
Complete prior elicitation dashboard for the CCT multisite trial (J = 38, μ_K = 5).
#> TableGrob (2 x 2) "dpprior_dashboard": 4 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) dpprior_dashboard gtable[layout]
#> 2 2 (2-2,1-1) dpprior_dashboard gtable[layout]
#> 3 3 (1-1,2-2) dpprior_dashboard gtable[layout]
#> 4 4 (2-2,2-2) dpprior_dashboard gtable[layout]1.6 Addressing the Dominance Problem
The diagnostics reveal an important concern: high dominance risk with . This means there is a 54% prior probability that a single cluster contains more than half of all sites.
In a setting with limited between-site information, this high dominance probability can lead to overshrinkage in posterior inference. The dual-anchor framework allows us to constrain the prior to reduce dominance while maintaining our beliefs about the expected number of clusters.
Applying dual-anchor refinement:
We target (25% dominance probability) while keeping as the primary anchor ():
# Apply dual-anchor to reduce dominance risk
fit_cct_dual <- DPprior_dual(
fit = fit_cct,
w1_target = list(prob = list(threshold = 0.5, value = 0.25)),
lambda = 0.7, # K remains primary anchor
loss_type = "adaptive",
verbose = FALSE
)
cat("Original prior: Gamma(a =", round(fit_cct$a, 4),
", b =", round(fit_cct$b, 4), ")\n")
#> Original prior: Gamma(a = 0.4629 , b = 0.2556 )
cat("Dual-anchor prior: Gamma(a =", round(fit_cct_dual$a, 4),
", b =", round(fit_cct_dual$b, 4), ")\n")
#> Dual-anchor prior: Gamma(a = 1.0887 , b = 0.4362 )Verifying the improvement:
diag_cct_dual <- DPprior_diagnostics(fit_cct_dual)
# Create comparison table
comparison_df <- data.frame(
Metric = c("E[α]", "E[K]", "Var(K)", "E[w₁]", "P(w₁ > 0.5)", "Dominance Risk"),
Original = c(
round(diag_cct$alpha$mean, 3),
round(diag_cct$K$mean, 2),
round(diag_cct$K$var, 2),
round(diag_cct$weights$mean, 3),
round(diag_cct$weights$prob_exceeds["prob_gt_0.5"], 3),
diag_cct$weights$dominance_risk
),
Dual_Anchor = c(
round(diag_cct_dual$alpha$mean, 3),
round(diag_cct_dual$K$mean, 2),
round(diag_cct_dual$K$var, 2),
round(diag_cct_dual$weights$mean, 3),
round(diag_cct_dual$weights$prob_exceeds["prob_gt_0.5"], 3),
diag_cct_dual$weights$dominance_risk
)
)
knitr::kable(
comparison_df,
col.names = c("Metric", "K-only Prior", "Dual-Anchor Prior"),
caption = "Comparison of K-only and dual-anchor priors for the CCT study"
)| Metric | K-only Prior | Dual-Anchor Prior |
|---|---|---|
| E[α] | 1.811 | 2.496 |
| E[K] | 5 | 6.65 |
| Var(K) | 20 | 18 |
| E[w₁] | 0.572 | 0.41 |
| P(w₁ > 0.5) | 0.545 | 0.355 |
| Dominance Risk | high | moderate |
1.7 Comparing K-only vs Dual-Anchor Priors
Let’s visualize the difference between the two priors:
# Prepare data for K PMF comparison
logS_cct <- compute_log_stirling(J_cct)
pmf_original <- pmf_K_marginal(J_cct, fit_cct$a, fit_cct$b, logS = logS_cct)
pmf_dual <- pmf_K_marginal(J_cct, fit_cct_dual$a, fit_cct_dual$b, logS = logS_cct)
k_compare_df <- data.frame(
K = rep(seq_along(pmf_original), 2),
Probability = c(pmf_original, pmf_dual),
Prior = rep(c("K-only", "Dual-anchor"), each = length(pmf_original))
)
k_compare_df$Prior <- factor(k_compare_df$Prior, levels = c("K-only", "Dual-anchor"))
# Prepare data for w1 comparison
w1_grid <- seq(0, 1, length.out = 200)
w1_dens_original <- sapply(w1_grid, function(w) density_w1(w, fit_cct$a, fit_cct$b))
w1_dens_dual <- sapply(w1_grid, function(w) density_w1(w, fit_cct_dual$a, fit_cct_dual$b))
w1_compare_df <- data.frame(
w1 = rep(w1_grid, 2),
Density = c(w1_dens_original, w1_dens_dual),
Prior = rep(c("K-only", "Dual-anchor"), each = length(w1_grid))
)
w1_compare_df$Prior <- factor(w1_compare_df$Prior, levels = c("K-only", "Dual-anchor"))
# Create plots
p1 <- ggplot(k_compare_df[k_compare_df$K <= 15, ],
aes(x = K, y = Probability, fill = Prior)) +
geom_col(position = "dodge", alpha = 0.8) +
scale_fill_manual(values = palette_2) +
labs(
x = expression(K[J]),
y = "Probability",
title = "Cluster Count Distribution"
) +
theme_minimal() +
theme(legend.position = "bottom", legend.title = element_blank())
p2 <- ggplot(w1_compare_df, aes(x = w1, y = Density, color = Prior)) +
geom_line(linewidth = 1) +
geom_vline(xintercept = 0.5, linetype = "dashed", color = "gray50") +
scale_color_manual(values = palette_2) +
labs(
x = expression(w[1]),
y = "Density",
title = "Largest Cluster Weight Distribution"
) +
theme_minimal() +
theme(legend.position = "bottom", legend.title = element_blank())
gridExtra::grid.arrange(p1, p2, ncol = 2)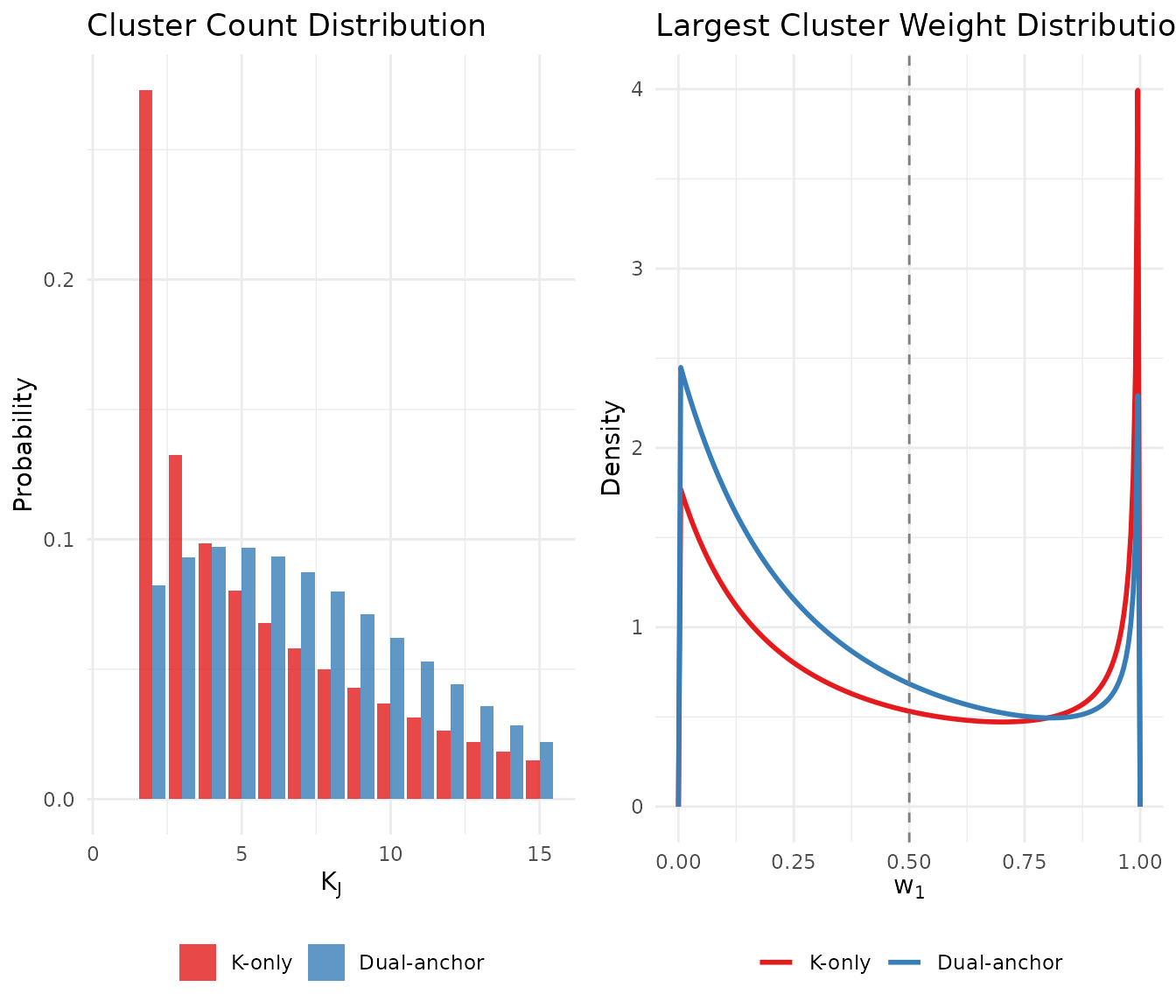
Comparison of K-only and dual-anchor priors: the dual-anchor approach reduces dominance risk while maintaining similar cluster count expectations.
The dual-anchor prior shifts mass in the distribution away from extreme values (right panel) while maintaining a similar expected number of clusters (left panel). This achieves our goal of reducing dominance risk without substantially changing our beliefs about cluster count.
1.8 Sensitivity Analysis
Given the uncertainty in our prior specification, we conduct a sensitivity analysis across a range of plausible values for both the K-only and dual-anchor approaches:
# Sensitivity to μ_K
mu_K_grid <- c(3, 5, 7, 10)
sensitivity_results <- lapply(mu_K_grid, function(mu) {
# K-only fit
fit_k <- DPprior_fit(J = J_cct, mu_K = mu, confidence = "low")
diag_k <- DPprior_diagnostics(fit_k)
# Dual-anchor fit
fit_d <- DPprior_dual(
fit = fit_k,
w1_target = list(prob = list(threshold = 0.5, value = 0.25)),
lambda = 0.7,
loss_type = "adaptive",
verbose = FALSE
)
diag_d <- DPprior_diagnostics(fit_d)
data.frame(
mu_K = mu,
# K-only results
a_k = round(fit_k$a, 3),
b_k = round(fit_k$b, 3),
E_K_k = round(diag_k$K$mean, 2),
P_dom_k = round(diag_k$weights$prob_exceeds["prob_gt_0.5"], 3),
# Dual-anchor results
a_d = round(fit_d$a, 3),
b_d = round(fit_d$b, 3),
E_K_d = round(diag_d$K$mean, 2),
P_dom_d = round(diag_d$weights$prob_exceeds["prob_gt_0.5"], 3)
)
})
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 74.3% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 54.5% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
sensitivity_df <- do.call(rbind, sensitivity_results)
knitr::kable(
sensitivity_df,
col.names = c("Target μ_K",
"a (K)", "b (K)", "E[K]", "P(w₁>0.5)",
"a (DA)", "b (DA)", "E[K]", "P(w₁>0.5)"),
row.names = FALSE,
caption = "Sensitivity analysis: K-only (K) vs Dual-anchor (DA) priors (J = 38, low confidence)"
)| Target μ_K | a (K) | b (K) | E[K] | P(w₁>0.5) | a (DA) | b (DA) | E[K] | P(w₁>0.5) |
|---|---|---|---|---|---|---|---|---|
| 3 | 0.270 | 0.345 | 3 | 0.743 | 7.568 | 3.733 | 6.45 | 0.276 |
| 5 | 0.463 | 0.256 | 5 | 0.545 | 1.089 | 0.436 | 6.65 | 0.355 |
| 7 | 0.601 | 0.191 | 7 | 0.399 | 0.840 | 0.232 | 7.94 | 0.313 |
| 10 | 0.727 | 0.123 | 10 | 0.253 | 0.732 | 0.124 | 10.03 | 0.251 |
1.9 Final Prior Selection and Reporting
Based on our analysis, we recommend the dual-anchor prior for the CCT study, as it provides appropriate control over dominance risk in this low between-site information setting.
cat("RECOMMENDED PRIOR FOR CCT STUDY\n")
#> RECOMMENDED PRIOR FOR CCT STUDY
cat("================================\n")
#> ================================
cat("α ~ Gamma(", round(fit_cct_dual$a, 4), ", ", round(fit_cct_dual$b, 4), ")\n\n", sep = "")
#> α ~ Gamma(1.0887, 0.4362)
cat("Key properties:\n")
#> Key properties:
cat(" E[K] =", round(diag_cct_dual$K$mean, 2), "clusters\n")
#> E[K] = 6.65 clusters
cat(" P(w₁ > 0.5) =", round(diag_cct_dual$weights$prob_exceeds["prob_gt_0.5"], 2), "\n")
#> P(w₁ > 0.5) = 0.36
cat(" Dominance risk:", diag_cct_dual$weights$dominance_risk, "\n")
#> Dominance risk: moderateReporting language for publication:
We elicited beliefs about the number of latent clusters among the sites in the CCT multisite trial. Based on substantive knowledge of similar education interventions and the uniformity of implementation, we specified (approximately five distinct response patterns) with low confidence. Initial diagnostics using the K-only calibration revealed high dominance risk with , which is problematic in settings with limited between-site information. We therefore applied dual-anchor refinement targeting with , yielding a Gamma(1.089, 0.436) hyperprior. The refined prior maintains while reducing dominance risk to moderate. Sensitivity analyses across confirmed that substantive conclusions were robust to prior specification.
2. Case Study: Brief Alcohol Interventions Meta-Analysis
2.1 Research Context
Our second case study is based on the meta-analysis of brief alcohol interventions (BAI) for adolescents and young adults conducted by Pustejovsky & Tipton (2022), who re-analyzed data from Tanner-Smith & Lipsey (2015).
Study characteristics:
- Number of studies: 117 randomized trials
- Total effect sizes: 1,198 estimates
- Effect sizes per study: Median = 6, range = 1 to 108
- Outcome: Alcohol consumption (standardized mean differences)
- Key feature: Complex dependence structure (correlated and hierarchical)
2.2 The Meta-Analytic Challenge
In meta-analysis, the Dirichlet Process mixture model can be used to flexibly model the distribution of true study effects. This is particularly valuable when:
- The true effect distribution may be non-Gaussian
- There may be discrete subpopulations of studies with similar effects
- Robust inference is needed without strong distributional assumptions
The BAI meta-analysis presents a setting with substantial between-study heterogeneity ( under the correlated effects model), suggesting the potential for distinct effect subgroups.
2.3 Eliciting the Prior
Step 1: Substantive reasoning about clusters
In the BAI context, we consider potential sources of effect heterogeneity:
- Intervention type: Brief motivational interviewing vs. feedback-only vs. multi-session programs vs. computerized interventions
- Population: College students vs. high school vs. clinical samples vs. community populations
- Outcome timing: Immediate vs. short-term (3 months) vs. medium-term (6 months) vs. long-term (12+ months) effects
- Control condition: No treatment vs. attention control vs. treatment-as-usual vs. alternative intervention
- Delivery format: Individual vs. group vs. self-administered
- Setting: Healthcare vs. educational vs. community
Given the many potential mechanisms for heterogeneity and the large number of studies, a reasonable expectation might be 15–25 distinct effect clusters among 117 studies. We start with :
# Define the meta-analysis context
J_bai <- 117
mu_K_bai <- 18
cat("Meta-analysis context:\n")
#> Meta-analysis context:
cat(" Number of studies (J):", J_bai, "\n")
#> Number of studies (J): 117
cat(" Expected clusters (μ_K):", mu_K_bai, "\n")
#> Expected clusters (μ_K): 18
cat(" Ratio (μ_K/J):", round(mu_K_bai / J_bai, 3), "\n")
#> Ratio (μ_K/J): 0.154Step 2: Expressing uncertainty
Meta-analyses often have more uncertainty about the number of distinct effect clusters than multisite trials, as studies are conducted independently with varying methodologies. We use low confidence:
fit_bai <- DPprior_fit(
J = J_bai,
mu_K = mu_K_bai,
confidence = "low"
)
print(fit_bai)
#> DPprior Prior Elicitation Result
#> =============================================
#>
#> Gamma Hyperprior: α ~ Gamma(a = 1.9005, b = 0.2986)
#> E[α] = 6.365, SD[α] = 4.617
#>
#> Target (J = 117):
#> E[K_J] = 18.00
#> Var(K_J) = 85.00
#> (from confidence = 'low')
#>
#> Achieved:
#> E[K_J] = 18.000000, Var(K_J) = 85.000000
#> Residual = 2.16e-12
#>
#> Method: A2-MN (6 iterations)
#>
#> Dominance Risk: LOW ✓ (P(w₁>0.5) = 10%)Step 3: Diagnostic verification
diag_bai <- DPprior_diagnostics(fit_bai)
cat("Diagnostic Summary:\n")
#> Diagnostic Summary:
cat("----------------\n")
#> ----------------
cat("Alpha distribution:\n")
#> Alpha distribution:
cat(" E[α] =", round(diag_bai$alpha$mean, 3), "\n")
#> E[α] = 6.365
cat(" SD(α) =", round(diag_bai$alpha$sd, 3), "\n")
#> SD(α) = 4.617
cat(" 95% CI: [", round(diag_bai$alpha$quantiles["q5"], 3), ", ",
round(diag_bai$alpha$quantiles["q95"], 3), "]\n", sep = "")
#> 95% CI: [1.059, 15.344]
cat("\nCluster count:\n")
#>
#> Cluster count:
cat(" E[K] =", round(diag_bai$K$mean, 3), "\n")
#> E[K] = 18
cat(" Var(K) =", round(diag_bai$K$var, 3), "\n")
#> Var(K) = 85
cat(" Mode(K) =", diag_bai$K$mode, "\n")
#> Mode(K) = 14
cat("\nWeight behavior:\n")
#>
#> Weight behavior:
cat(" E[w₁] =", round(diag_bai$weights$mean, 3), "\n")
#> E[w₁] = 0.2
cat(" P(w₁ > 0.5) =",
round(diag_bai$weights$prob_exceeds["prob_gt_0.5"], 3), "\n")
#> P(w₁ > 0.5) = 0.102
cat(" Dominance risk:", diag_bai$weights$dominance_risk, "\n")
#> Dominance risk: low2.4 Comparing Alternative Specifications
For meta-analysis, it is valuable to compare different prior specifications to assess robustness. We compare three approaches: expecting fewer clusters, the baseline, and expecting more clusters.
# Define three candidate priors
bai_candidates <- list(
"Conservative (μ_K = 10)" = DPprior_fit(J = J_bai, mu_K = 10, confidence = "low"),
"Baseline (μ_K = 18)" = DPprior_fit(J = J_bai, mu_K = 18, confidence = "low"),
"Liberal (μ_K = 30)" = DPprior_fit(J = J_bai, mu_K = 30, confidence = "low")
)
# Create comparison table
bai_comp_results <- lapply(names(bai_candidates), function(nm) {
fit <- bai_candidates[[nm]]
diag <- DPprior_diagnostics(fit)
data.frame(
Prior = nm,
a = round(fit$a, 3),
b = round(fit$b, 3),
E_K = round(diag$K$mean, 2),
E_w1 = round(diag$weights$mean, 3),
E_rho = round(diag$coclustering$mean, 3)
)
})
bai_comp_df <- do.call(rbind, bai_comp_results)
knitr::kable(
bai_comp_df,
col.names = c("Prior", "a", "b", "E[K]", "E[w₁]", "E[ρ]"),
caption = "Comparison of candidate priors for the BAI meta-analysis (J = 117)"
)| Prior | a | b | E[K] | E[w₁] | E[ρ] |
|---|---|---|---|---|---|
| Conservative (μ_K = 10) | 1.232 | 0.449 | 10 | 0.379 | 0.379 |
| Baseline (μ_K = 18) | 1.901 | 0.299 | 18 | 0.200 | 0.200 |
| Liberal (μ_K = 30) | 2.470 | 0.173 | 30 | 0.096 | 0.096 |
2.5 Visualizing the Comparison
# Create data for K PMF comparison
logS_bai <- compute_log_stirling(J_bai)
k_bai_df <- do.call(rbind, lapply(names(bai_candidates), function(nm) {
fit <- bai_candidates[[nm]]
pmf <- pmf_K_marginal(J_bai, fit$a, fit$b, logS = logS_bai)
data.frame(
K = seq_along(pmf),
probability = pmf,
Prior = nm
)
}))
k_bai_df$Prior <- factor(k_bai_df$Prior, levels = names(bai_candidates))
# Plot PMF comparison
ggplot(k_bai_df[k_bai_df$K <= 50, ],
aes(x = K, y = probability, color = Prior)) +
geom_line(linewidth = 0.8) +
geom_point(size = 1.5) +
scale_color_manual(values = palette_3) +
labs(
x = expression(K[J]),
y = "Probability",
title = "Prior PMF of Number of Clusters (BAI Meta-Analysis)",
subtitle = "J = 117 studies"
) +
theme_minimal() +
theme(legend.position = "bottom", legend.title = element_blank())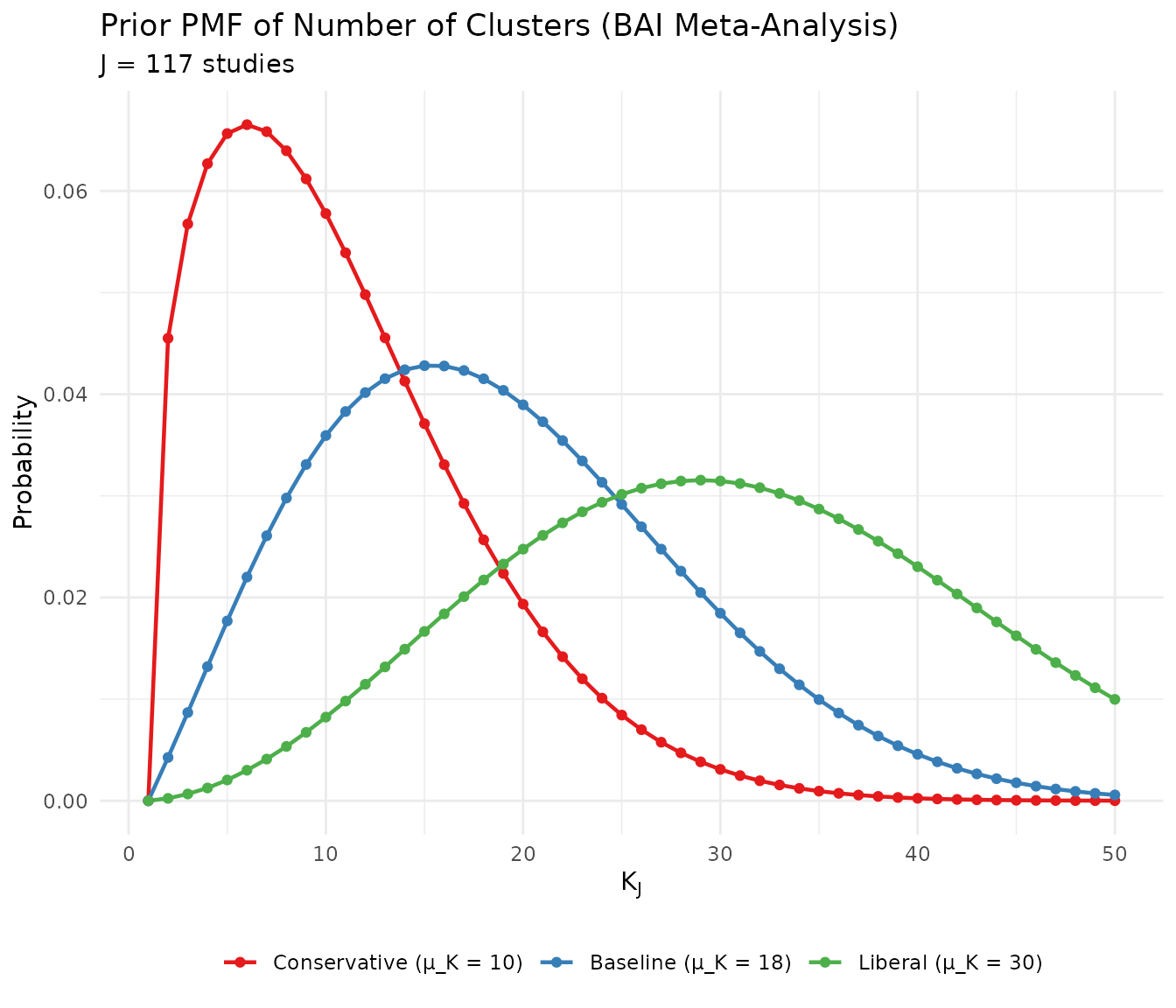
Comparison of three candidate priors for the BAI meta-analysis.
3. Case Study: Alabama Pre-K Value-Added Analysis
3.1 Research Context
Our third case study illustrates prior elicitation for a large-scale value-added analysis in Alabama’s pre-kindergarten program.
Study characteristics:
- Sites: 847 pre-K sites across the state
- Providers: 74.4% public schools, 10.4% private centers, 6.2% Head Start, and others
- Class sizes: 13–18 students per classroom
- Outcomes: Six developmental domains (Literacy, Mathematics, Cognitive, Language, Social-Emotional, Physical)
- Assessment: Teaching Strategies GOLD®
A note on package limitations: The current version of DPprior supports due to pre-computed unsigned Stirling numbers of the first kind, which are required for exact moment calculations. Support for larger is planned for future releases. For the 847 sites in the Alabama study, we demonstrate the workflow using as a practical upper bound. This limitation has minimal impact on substantive conclusions for several reasons: (1) the relationship between and is approximately logarithmic in , so results are relatively stable across nearby sample sizes; (2) using a conservative (smaller) tends to produce slightly more concentrated priors, which is a defensible choice in uncertain settings; and (3) the ratio—rather than alone—is the primary driver of the elicited prior.
3.2 The Value-Added Challenge
Value-added models (VAMs) estimate site-specific contributions to student learning (). Key challenges in this context include:
- Distribution estimation: Is the distribution of site effects normal, or are there distinct subpopulations?
- Tail identification: Which sites fall in the lowest and highest 10% of the distribution?
- Ranking: Can we produce meaningful “report cards” for sites?
The semiparametric Dirichlet Process approach allows flexible modeling of the site effect distribution without imposing normality.
3.3 Eliciting the Prior
Step 1: Substantive considerations
With 847 sites, we expect considerable heterogeneity. Potential sources of clustering include:
- Provider type: Public schools vs. private centers vs. Head Start may exhibit systematically different effects
- Geographic region: Urban vs. rural patterns
- Resource levels: Varying levels of site resources and teacher qualifications
- Implementation fidelity: Variation in how the curriculum is delivered
A reasonable expectation might be 40–60 distinct “types” of sites, representing approximately 5–7% of the total. We specify :
# Define the context
# Note: Using J = 500 (package maximum) for the 847-site study
J_alabama <- 500
mu_K_alabama <- 50
cat("Alabama Pre-K context:\n")
#> Alabama Pre-K context:
cat(" Actual number of sites: 847\n")
#> Actual number of sites: 847
cat(" J used for elicitation:", J_alabama, "(package maximum)\n")
#> J used for elicitation: 500 (package maximum)
cat(" Expected clusters (μ_K):", mu_K_alabama, "\n")
#> Expected clusters (μ_K): 50
cat(" Ratio (μ_K/J):", round(mu_K_alabama / J_alabama, 3), "\n")
#> Ratio (μ_K/J): 0.1Step 2: Prior elicitation
# Fit using A2-MN method
fit_alabama <- DPprior_fit(
J = J_alabama,
mu_K = mu_K_alabama,
confidence = "low",
method = "A2-MN"
)
print(fit_alabama)
#> DPprior Prior Elicitation Result
#> =============================================
#>
#> Gamma Hyperprior: α ~ Gamma(a = 6.2200, b = 0.4431)
#> E[α] = 14.038, SD[α] = 5.629
#>
#> Target (J = 500):
#> E[K_J] = 50.00
#> Var(K_J) = 245.00
#> (from confidence = 'low')
#>
#> Achieved:
#> E[K_J] = 50.000000, Var(K_J) = 245.000000
#> Residual = 2.84e-14
#>
#> Method: A2-MN (6 iterations)
#>
#> Dominance Risk: LOW ✓ (P(w₁>0.5) = 0%)Step 3: Diagnostics
diag_alabama <- DPprior_diagnostics(fit_alabama)
cat("Diagnostic Summary:\n")
#> Diagnostic Summary:
cat("----------------\n")
#> ----------------
cat("Alpha distribution:\n")
#> Alpha distribution:
cat(" E[α] =", round(diag_alabama$alpha$mean, 3), "\n")
#> E[α] = 14.038
cat(" SD(α) =", round(diag_alabama$alpha$sd, 3), "\n")
#> SD(α) = 5.629
cat(" 95% CI: [", round(diag_alabama$alpha$quantiles["q5"], 3), ", ",
round(diag_alabama$alpha$quantiles["q95"], 3), "]\n", sep = "")
#> 95% CI: [6.226, 24.392]
cat("\nCluster count:\n")
#>
#> Cluster count:
cat(" E[K] =", round(diag_alabama$K$mean, 2), "\n")
#> E[K] = 50
cat(" SD(K) =", round(sqrt(diag_alabama$K$var), 2), "\n")
#> SD(K) = 15.65
cat("\nWeight behavior:\n")
#>
#> Weight behavior:
cat(" E[w₁] =", round(diag_alabama$weights$mean, 4), "\n")
#> E[w₁] = 0.077
cat(" P(w₁ > 0.5) =",
round(diag_alabama$weights$prob_exceeds["prob_gt_0.5"], 4), "\n")
#> P(w₁ > 0.5) = 0.0029
cat(" Dominance risk:", diag_alabama$weights$dominance_risk, "\n")
#> Dominance risk: low3.4 Sensitivity Analysis
We examine how the elicited prior changes across different specifications:
# Sensitivity to μ_K
mu_K_grid <- c(30, 40, 50, 60, 75)
sensitivity_alabama <- lapply(mu_K_grid, function(mu) {
fit <- DPprior_fit(J = J_alabama, mu_K = mu, confidence = "low", method = "A2-MN")
diag <- DPprior_diagnostics(fit)
data.frame(
mu_K = mu,
ratio = round(mu / J_alabama, 3),
a = round(fit$a, 3),
b = round(fit$b, 3),
E_K = round(diag$K$mean, 1),
P_dom = round(diag$weights$prob_exceeds["prob_gt_0.5"], 4)
)
})
sensitivity_alabama_df <- do.call(rbind, sensitivity_alabama)
knitr::kable(
sensitivity_alabama_df,
col.names = c("Target μ_K", "μ_K/J", "a", "b", "E[K]", "P(w₁ > 0.5)"),
caption = "Sensitivity analysis for the Alabama Pre-K study (J = 500)"
)| Target μ_K | μ_K/J | a | b | E[K] | P(w₁ > 0.5) | |
|---|---|---|---|---|---|---|
| prob_gt_0.5 | 30 | 0.06 | 4.171 | 0.588 | 30 | 0.0388 |
| prob_gt_0.51 | 40 | 0.08 | 5.252 | 0.506 | 40 | 0.0108 |
| prob_gt_0.52 | 50 | 0.10 | 6.220 | 0.443 | 50 | 0.0029 |
| prob_gt_0.53 | 60 | 0.12 | 7.087 | 0.392 | 60 | 0.0007 |
| prob_gt_0.54 | 75 | 0.15 | 8.215 | 0.330 | 75 | 0.0001 |
3.5 Visualizing Prior Behavior
# Plot alpha and K distributions side by side
# Alpha distribution
alpha_grid <- seq(0.01, 25, length.out = 300)
alpha_df <- data.frame(
alpha = alpha_grid,
density = dgamma(alpha_grid, shape = fit_alabama$a, rate = fit_alabama$b)
)
p1 <- ggplot(alpha_df, aes(x = alpha, y = density)) +
geom_line(linewidth = 1, color = "#377EB8") +
geom_vline(xintercept = diag_alabama$alpha$mean,
linetype = "dashed", color = "gray40") +
annotate("text", x = diag_alabama$alpha$mean + 1.5, y = max(alpha_df$density) * 0.9,
label = sprintf("E[α] = %.1f", diag_alabama$alpha$mean),
hjust = 0, color = "gray40") +
labs(
x = expression(alpha),
y = "Density",
title = expression("Prior on " * alpha)
) +
theme_minimal()
# K distribution
logS_al <- compute_log_stirling(J_alabama)
pmf_K <- pmf_K_marginal(J_alabama, fit_alabama$a, fit_alabama$b, logS = logS_al)
k_df <- data.frame(
K = seq_along(pmf_K),
probability = pmf_K
)
p2 <- ggplot(k_df[k_df$K <= 100 & k_df$probability > 1e-4, ],
aes(x = K, y = probability)) +
geom_col(fill = "#377EB8", alpha = 0.7) +
geom_vline(xintercept = diag_alabama$K$mean,
linetype = "dashed", color = "gray40") +
annotate("text", x = diag_alabama$K$mean + 5, y = max(k_df$probability) * 0.9,
label = sprintf("E[K] = %.0f", diag_alabama$K$mean),
hjust = 0, color = "gray40") +
labs(
x = expression(K[J]),
y = "Probability",
title = expression("Prior PMF of " * K[J])
) +
theme_minimal()
gridExtra::grid.arrange(p1, p2, ncol = 2)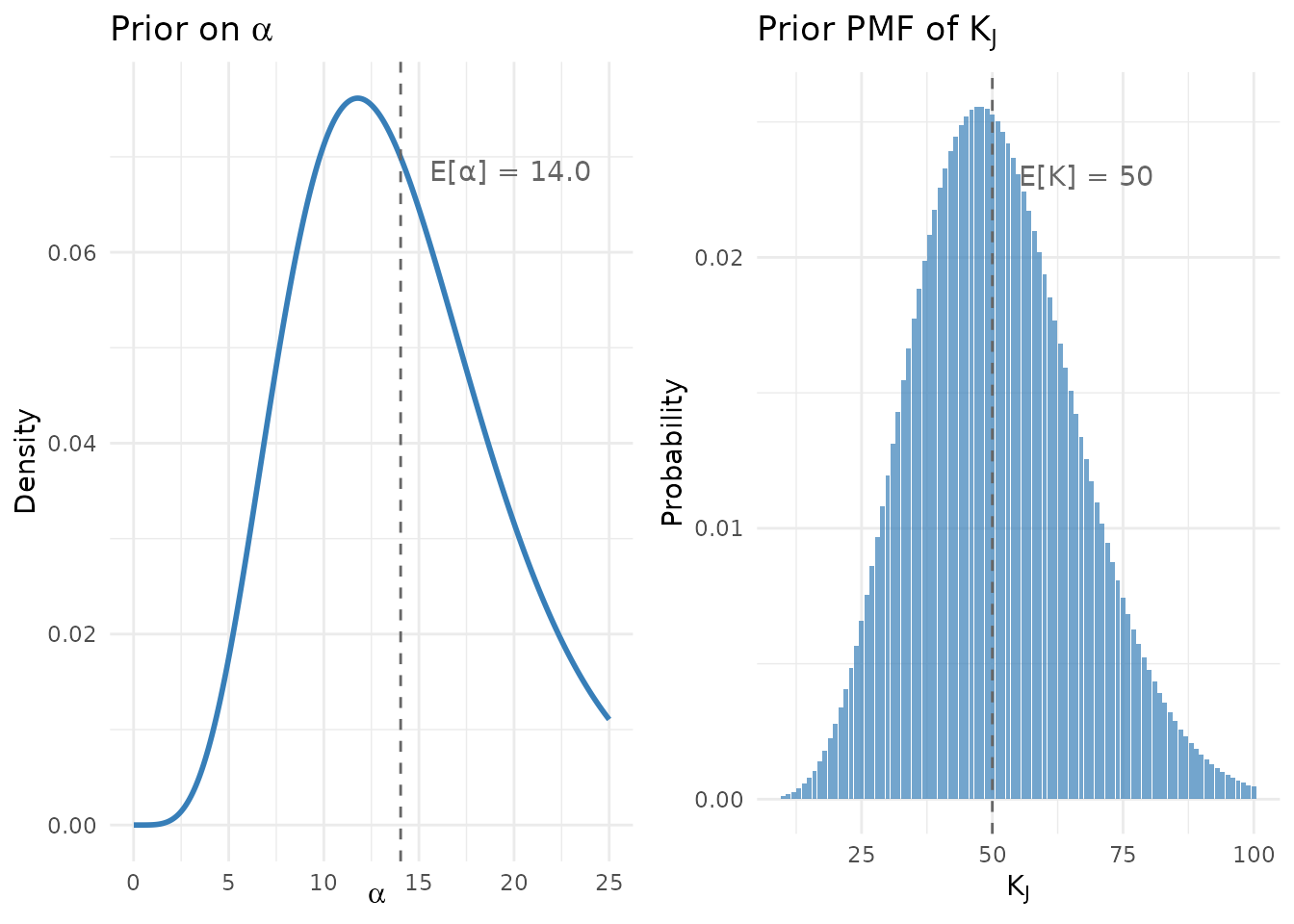
Prior distributions for the Alabama Pre-K study (J = 500, μ_K = 50).
4. Reporting Prior Elicitation in Publications
4.1 Key Elements to Report
When reporting prior elicitation in a publication, include:
- Study context: Number of units () and their nature
- Substantive rationale: Why you expect a certain number of clusters
- Uncertainty characterization: Confidence level or variance specification
- Algorithm used: A1, A2-MN, or A2-KL
- Resulting hyperparameters: The specification
- Diagnostic summary: Key quantities like and dominance risk
- Sensitivity analysis: Results for alternative specifications
Summary
This vignette demonstrated how to apply the DPprior package across three diverse research contexts:
| Case Study | J | μ_K | Key Insight |
|---|---|---|---|
| CCT Multisite Trial | 38 | 5 | Dual-anchor refinement essential for low-information settings |
| BAI Meta-Analysis | 117 | 18 | Multiple heterogeneity sources suggest more clusters |
| Alabama Pre-K VAM | 500 | 50 | Large-scale applications work within package limits |
Key Takeaways:
Context matters: The appropriate depends on substantive domain knowledge about heterogeneity sources
Diagnostics are essential: Always verify weight behavior, not just cluster counts
Dual-anchor refinement: When dominance risk is high, use
DPprior_dual()to control weight behaviorSensitivity analysis is mandatory: Report results across plausible alternative specifications
Transparent reporting: Document the complete elicitation process in publications
References
Barrera-Osorio, F., Linden, L. L., & Saavedra, J. E. (2019). Medium- and long-term educational consequences of alternative conditional cash transfer designs: Experimental evidence from Colombia. American Economic Journal: Applied Economics, 11(3), 54–91. https://doi.org/10.1257/app.20170008
Lee, J., Che, J., Rabe-Hesketh, S., Feller, A., & Miratrix, L. (2025). Improving the estimation of site-specific effects and their distribution in multisite trials. Journal of Educational and Behavioral Statistics, 50(5), 731–764. https://doi.org/10.3102/10769986241254286
Pustejovsky, J. E., & Tipton, E. (2022). Meta-analysis with robust variance estimation: Expanding the range of working models. Prevention Science, 23(3), 425–438. https://doi.org/10.1007/s11121-021-01246-3
Tanner-Smith, E. E., & Lipsey, M. W. (2015). Brief alcohol interventions for adolescents and young adults: A systematic review and meta-analysis. Journal of Substance Abuse Treatment, 51, 1–18. https://doi.org/10.1016/j.jsat.2014.09.001
For questions or feedback, please visit the GitHub repository.