From Poisson Proxy to NegBin: The A1 Approximation Theory
JoonHo Lee
2026-01-01
Source:vignettes/theory-approximations.Rmd
theory-approximations.RmdOverview
This vignette provides a rigorous mathematical treatment of the A1 closed-form approximation used in the DPprior package for Gamma hyperprior elicitation. The A1 method transforms practitioner beliefs about the number of clusters into Gamma hyperparameters via a Negative Binomial moment-matching procedure.
We cover:
- The Poisson approximation for large and its theoretical foundations
- Scaling constant selection: vs harmonic vs digamma
- The Negative Binomial gamma mixture identity
- The A1 inverse mapping (Theorem 1)
- Error analysis and limitations of the approximation
Throughout, we carefully distinguish between established results from the literature and novel contributions of this work.
1. The Poisson Approximation for Large J
1.1 Established Result: Poisson Convergence
The starting point for the A1 approximation is a classical result on
the asymptotic distribution of
.
Recall from the Poisson-Binomial representation (see
vignette("theory-overview")) that:
Attribution. The following Poisson convergence result is established in Arratia, Barbour, and Tavaré (2000), with the rate explicitly stated in Vicentini and Jermyn (2025, Equation 10):
Theorem (Poisson Approximation). For fixed and : where denotes the total variation distance.
This convergence, while sublinear, provides the conceptual foundation for using a Poisson proxy in elicitation.
1.2 The Shifted Poisson Proxy
Novel Contribution. A key insight motivating the DPprior approach is that we should apply the Poisson approximation to rather than directly.
Rationale. Any unshifted Poisson approximation has an unavoidable support mismatch because almost surely under the CRP (at least one cluster always exists). If , then:
For small (which occurs when the elicited is close to 1), this creates a substantial lower bound on the approximation error.
The A1 proxy (shifted): or equivalently .
This shift aligns naturally with Proposition 1 from the theory vignette, since is a sum of independent Bernoulli variables ().
# Visualize the CRP probabilities
J <- 50
alpha_values <- c(0.5, 1, 2, 5)
crp_data <- do.call(rbind, lapply(alpha_values, function(a) {
i <- 1:J
p_new <- a / (a + i - 1)
data.frame(
customer = i,
prob_new_table = p_new,
alpha = paste0("α = ", a)
)
}))
crp_data$alpha <- factor(crp_data$alpha, levels = paste0("α = ", alpha_values))
ggplot(crp_data, aes(x = customer, y = prob_new_table, color = alpha)) +
geom_line(linewidth = 1) +
scale_color_manual(values = palette_main[1:4]) +
labs(
x = "Customer i",
y = expression("P(new table) = " * alpha / (alpha + i - 1)),
title = "CRP: Probability of Starting a New Table",
subtitle = "The expected number of tables ≈ α · log(J)"
) +
theme_minimal() +
theme(legend.position = "right")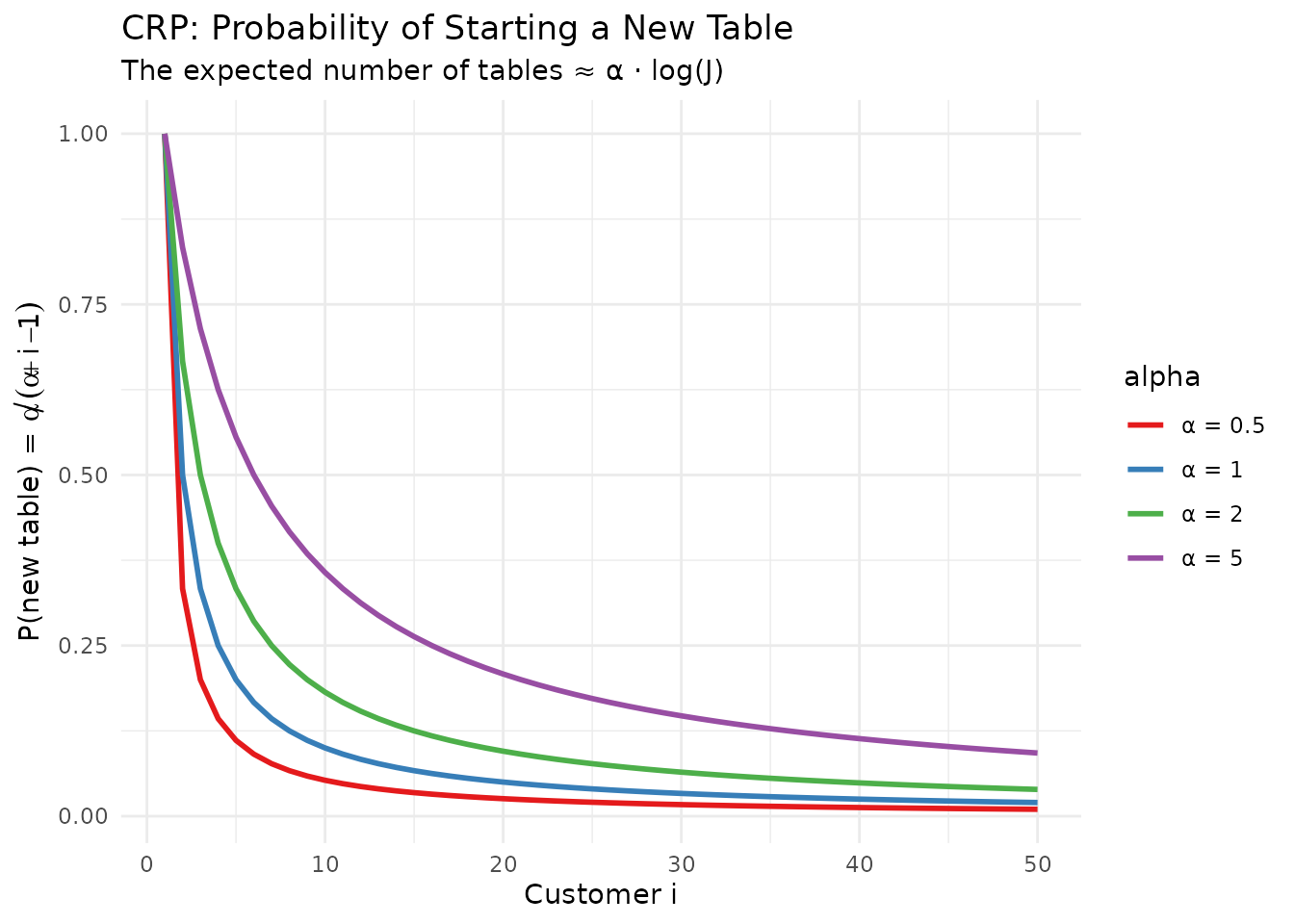
Intuition: The probability of opening a new table decreases as customers arrive.
1.3 Mean Approximation
The exact conditional mean is (see
vignette("theory-overview")):
For the shifted mean: where is a scaling constant that we now analyze.
2. The Scaling Constant Family
2.1 Three Candidates
The A1 approximation requires a deterministic scaling constant satisfying . Three natural candidates emerge from asymptotic considerations:
| Variant | Formula | Description |
|---|---|---|
| log (default) | Asymptotic leading term | |
| harmonic | Better for moderate | |
| digamma | Local correction |
where is the Euler-Mascheroni constant and is a plug-in estimate.
Lemma (Harmonic-Digamma Equivalence). For integer :
Thus, there are only two distinct mean-level competitors for integer : and .
# Compare scaling constants across J values
J_values <- c(10, 25, 50, 100, 200, 500)
scaling_df <- data.frame(
J = J_values,
log_J = sapply(J_values, function(J) compute_scaling_constant(J, "log")),
harmonic = sapply(J_values, function(J) compute_scaling_constant(J, "harmonic"))
)
scaling_df$difference <- scaling_df$harmonic - scaling_df$log_J
scaling_df$ratio <- scaling_df$harmonic / scaling_df$log_J
knitr::kable(
scaling_df,
digits = 4,
col.names = c("J", "log(J)", "H_{J-1}", "Difference", "Ratio"),
caption = "Comparison of scaling constants"
)| J | log(J) | H_{J-1} | Difference | Ratio |
|---|---|---|---|---|
| 10 | 2.3026 | 2.8290 | 0.5264 | 1.2286 |
| 25 | 3.2189 | 3.7760 | 0.5571 | 1.1731 |
| 50 | 3.9120 | 4.4792 | 0.5672 | 1.1450 |
| 100 | 4.6052 | 5.1774 | 0.5722 | 1.1243 |
| 200 | 5.2983 | 5.8730 | 0.5747 | 1.1085 |
| 500 | 6.2146 | 6.7908 | 0.5762 | 1.0927 |
2.2 Asymptotic Analysis
Attribution. The following asymptotic expansion is standard (see Abramowitz & Stegun, 1964):
Proposition (Large-J Expansion). For fixed and :
Moreover:
Key insight: The “best” -free linear approximation cannot be uniform in because the second-order term depends on .
# Show asymptotic behavior
J <- 100
alpha_grid <- seq(0.1, 5, length.out = 100)
asymp_data <- data.frame(
alpha = alpha_grid,
exact = mean_K_given_alpha(J, alpha_grid),
log_approx = alpha_grid * log(J) + 1,
harmonic_approx = alpha_grid * (digamma(J) + 0.5772156649) + 1
)
# Reshape for plotting
asymp_long <- data.frame(
alpha = rep(alpha_grid, 3),
mean_K = c(asymp_data$exact, asymp_data$log_approx, asymp_data$harmonic_approx),
Method = rep(c("Exact", "log(J) + 1", "H_{J-1} + 1"), each = length(alpha_grid))
)
asymp_long$Method <- factor(asymp_long$Method,
levels = c("Exact", "log(J) + 1", "H_{J-1} + 1"))
ggplot(asymp_long, aes(x = alpha, y = mean_K, color = Method, linetype = Method)) +
geom_line(linewidth = 1) +
scale_color_manual(values = c("#000000", "#E41A1C", "#377EB8")) +
scale_linetype_manual(values = c("solid", "dashed", "dotted")) +
labs(
x = expression(alpha),
y = expression(E[K[J] * " | " * alpha]),
title = expression("Exact Mean vs. Linear Approximations (J = 100)"),
subtitle = "The approximations diverge for small and large α"
) +
theme_minimal() +
theme(legend.position = "right")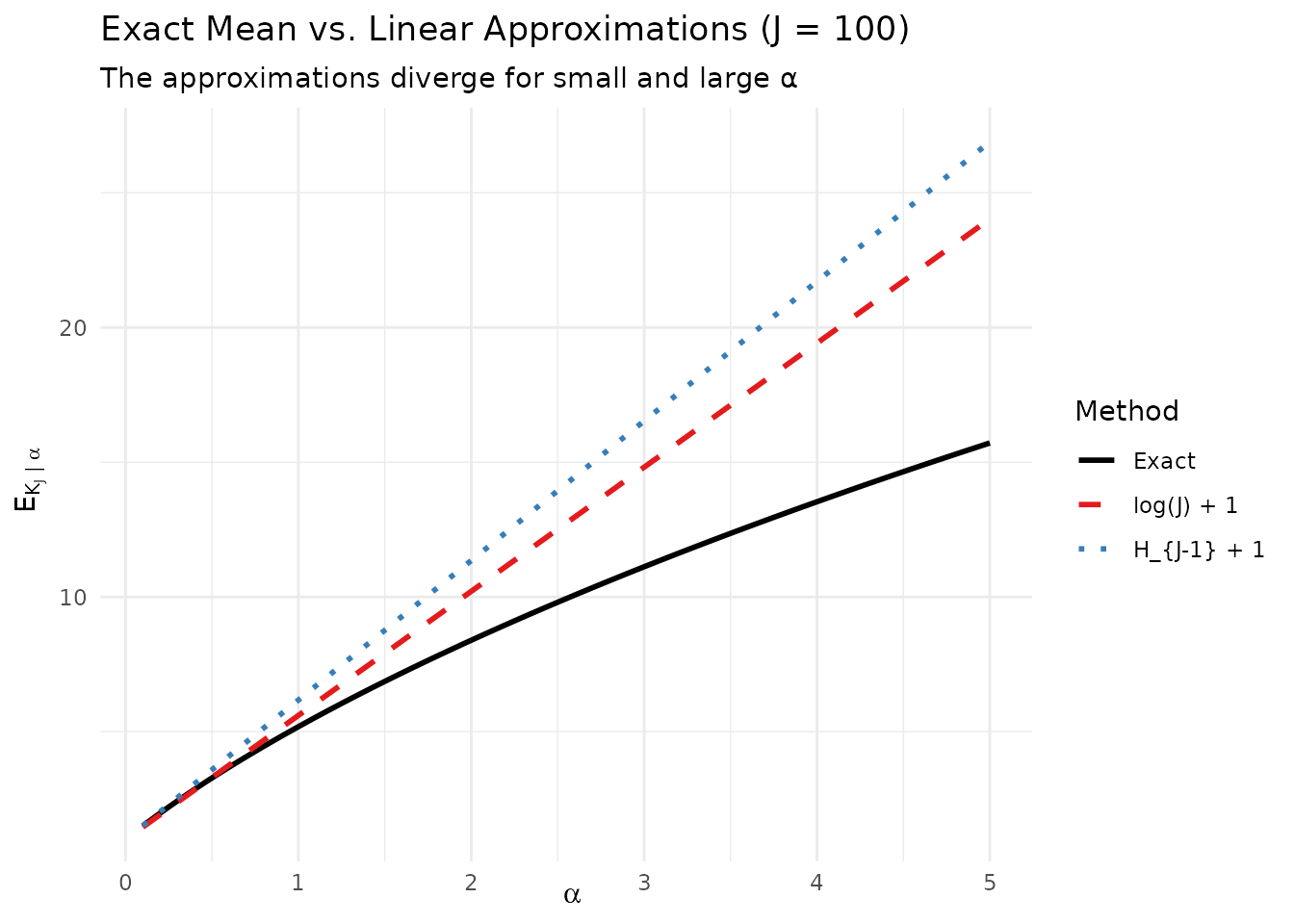
The exact mean μ_J(α) and its linear approximations.
2.3 Crossover Analysis
Novel Contribution. Define the crossover point as the value where the shifted mean errors are equal:
Proposition (Limiting Crossover). As : where solves .
Practical implication: For the shifted mean with (which covers essentially all practical applications), the scaling dominates in terms of mean approximation error.
# Compute mean errors for different scaling choices
J_test <- 50
alpha_grid <- seq(0.1, 3, by = 0.1)
error_df <- data.frame(
alpha = alpha_grid,
error_log = abs(mean_K_given_alpha(J_test, alpha_grid) - 1 -
alpha_grid * compute_scaling_constant(J_test, "log")),
error_harmonic = abs(mean_K_given_alpha(J_test, alpha_grid) - 1 -
alpha_grid * compute_scaling_constant(J_test, "harmonic"))
)
error_df$better <- ifelse(error_df$error_log < error_df$error_harmonic,
"log(J)", "H_{J-1}")
cat("Mean error comparison for J =", J_test, "\n")
#> Mean error comparison for J = 50
cat("α range where log(J) is better:",
sum(error_df$better == "log(J)"), "out of", nrow(error_df), "values\n")
#> α range where log(J) is better: 29 out of 30 values
cat("Crossover occurs near α ≈",
error_df$alpha[which.min(abs(error_df$error_log - error_df$error_harmonic))], "\n")
#> Crossover occurs near α ≈ 0.23. The NegBin Gamma Mixture
3.1 The Key Identity
Attribution. The following is a standard result in Bayesian statistics (see, e.g., the Gamma-Poisson conjugacy):
Proposition (Gamma-Poisson Mixture). If and , then:
More generally, if and , then:
3.2 Application to
Novel Application. Applying this to the A1 proxy with :
Under the NegBin parameterization where is the success probability:
| Quantity | Formula |
|---|---|
| Mean of | |
| Variance of | |
| Mean of | |
| Variance of |
# Compare NegBin approximation to exact marginal
J <- 50
a <- 1.5
b <- 0.5
logS <- compute_log_stirling(J)
# Exact marginal PMF
pmf_exact <- pmf_K_marginal(J, a, b, logS)
# NegBin approximation
c_J <- log(J)
p_nb <- b / (b + c_J)
# NegBin PMF for K-1, then shift
k_vals <- 0:J
pmf_negbin <- dnbinom(k_vals - 1, size = a, prob = p_nb)
pmf_negbin[1] <- 0 # K >= 1
# Normalize for comparison
pmf_negbin <- pmf_negbin / sum(pmf_negbin)
# Create comparison data
k_show <- 1:25
comparison_df <- data.frame(
K = rep(k_show, 2),
Probability = c(pmf_exact[k_show + 1], pmf_negbin[k_show + 1]),
Method = rep(c("Exact Marginal", "NegBin Approximation"), each = length(k_show))
)
ggplot(comparison_df, aes(x = K, y = Probability, fill = Method)) +
geom_bar(stat = "identity", position = "dodge", alpha = 0.8) +
scale_fill_manual(values = c("#377EB8", "#E41A1C")) +
labs(
x = expression(K[J]),
y = "Probability",
title = expression("NegBin Approximation vs Exact Marginal (J = 50)"),
subtitle = paste0("Gamma(", a, ", ", b, ") prior on α")
) +
theme_minimal() +
theme(legend.position = "bottom")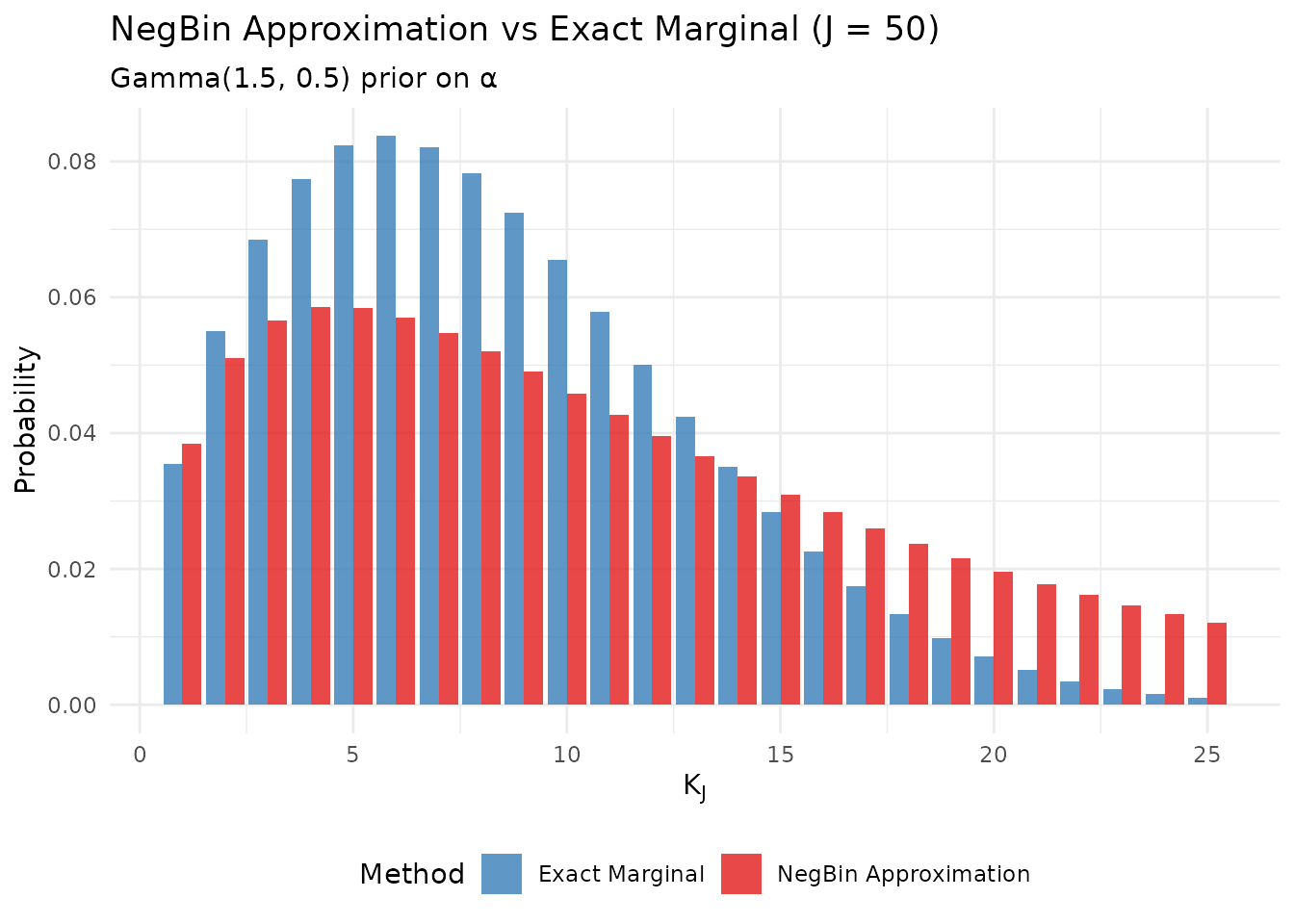
The NegBin approximation vs exact marginal distribution.
4. The A1 Inverse Mapping (Theorem 1)
4.1 The Inverse Problem
Given practitioner-specified beliefs about the number of clusters, we seek the Gamma hyperparameters such that the marginal distribution of has the desired moments.
Under the NegBin proxy, this becomes a moment-matching problem with an analytical solution.
4.2 Main Result
Novel Contribution. The following theorem provides the closed-form inverse mapping that is central to the A1 method:
Theorem 1 (A1 Inverse Mapping). Fix and scaling constant . Define the shifted mean .
If and (overdispersion), then there exists a unique that matches under the NegBin proxy, given by:
Proof. From the NegBin moments:
Substituting into the variance equation :
Solving for :
Positivity requires , which is the overdispersion condition.
4.3 Corollary: Interpretation for
Corollary 1. Under Theorem 1:
Interpretation: Under A1, determines the prior mean of , while the “confidence” (through ) determines the prior variance of .
# Demonstrate the A1 mapping
J <- 50
mu_K <- 5
var_K <- 8
# Step-by-step calculation
m <- mu_K - 1
D <- var_K - m
c_J <- log(J)
cat("A1 Inverse Mapping Demonstration\n")
#> A1 Inverse Mapping Demonstration
cat(paste(rep("=", 50), collapse = ""), "\n\n")
#> ==================================================
cat("Inputs:\n")
#> Inputs:
cat(sprintf(" J = %d\n", J))
#> J = 50
cat(sprintf(" μ_K = %.1f\n", mu_K))
#> μ_K = 5.0
cat(sprintf(" σ²_K = %.1f\n\n", var_K))
#> σ²_K = 8.0
cat("Intermediate calculations:\n")
#> Intermediate calculations:
cat(sprintf(" m = μ_K - 1 = %.1f\n", m))
#> m = μ_K - 1 = 4.0
cat(sprintf(" D = σ²_K - m = %.1f\n", D))
#> D = σ²_K - m = 4.0
cat(sprintf(" c_J = log(J) = %.4f\n\n", c_J))
#> c_J = log(J) = 3.9120
cat("A1 formulas:\n")
#> A1 formulas:
cat(sprintf(" a = m² / D = %.1f² / %.1f = %.4f\n", m, D, m^2/D))
#> a = m² / D = 4.0² / 4.0 = 4.0000
cat(sprintf(" b = m · c_J / D = %.1f × %.4f / %.1f = %.4f\n\n", m, c_J, D, m*c_J/D))
#> b = m · c_J / D = 4.0 × 3.9120 / 4.0 = 3.9120
# Use DPprior_a1 to verify
fit <- DPprior_a1(J = J, mu_K = mu_K, var_K = var_K)
cat("DPprior_a1() result:\n")
#> DPprior_a1() result:
cat(sprintf(" a = %.4f\n", fit$a))
#> a = 4.0000
cat(sprintf(" b = %.4f\n", fit$b))
#> b = 3.9120
cat(sprintf(" E[α] = a/b = %.4f\n", fit$a / fit$b))
#> E[α] = a/b = 1.0225
cat(sprintf(" CV(α) = 1/√a = %.4f\n", 1/sqrt(fit$a)))
#> CV(α) = 1/√a = 0.50004.4 Feasibility Region
Novel Contribution. The feasibility constraints for A1 are:
Shifted A1 (default, for DP partitions):
These are proxy-model constraints arising from the NegBin moment identity, not fundamental constraints of the DP itself.
Important: Some user specifications with may be feasible under the exact DP but infeasible under A1. In such cases, the package projects to the feasibility boundary.
# Demonstrate feasibility projection
cat("Feasibility Demonstration\n")
#> Feasibility Demonstration
cat(paste(rep("=", 50), collapse = ""), "\n\n")
#> ==================================================
# Case 1: Feasible
fit1 <- DPprior_a1(J = 50, mu_K = 5, var_K = 8)
cat("Case 1: Feasible (var_K = 8 > μ_K - 1 = 4)\n")
#> Case 1: Feasible (var_K = 8 > μ_K - 1 = 4)
cat(sprintf(" Status: %s\n\n", fit1$status))
#> Status: success
# Case 2: Infeasible - requires projection
fit2 <- DPprior_a1(J = 50, mu_K = 5, var_K = 3)
#> Warning: var_K <= mu_K - 1: projected to feasible boundary
cat("Case 2: Infeasible (var_K = 3 < μ_K - 1 = 4)\n")
#> Case 2: Infeasible (var_K = 3 < μ_K - 1 = 4)
cat(sprintf(" Status: %s\n", fit2$status))
#> Status: projected
cat(sprintf(" Original var_K: %.1f\n", 3))
#> Original var_K: 3.0
cat(sprintf(" Projected var_K: %.6f\n", fit2$target$var_K_used))5. Error Analysis
5.1 Sources of Approximation Error
The A1 method introduces several sources of error:
- Poisson proxy error: The finite- deviation from the Poisson limit
- Scaling constant choice: Different options introduce different biases
- Moment matching vs. distribution matching: Even if moments match, the full distributions may differ
5.2 Quantifying Moment Error
Novel Contribution. We can directly compare the A1-achieved moments against the exact marginal moments:
# Compare A1 moments to exact moments
test_cases <- expand.grid(
J = c(25, 50, 100, 200),
mu_K = c(5, 10),
vif = c(1.5, 2, 3)
)
# Filter valid cases first
test_cases <- test_cases[test_cases$mu_K < test_cases$J, ]
results <- do.call(rbind, lapply(1:nrow(test_cases), function(i) {
J <- test_cases$J[i]
mu_K <- test_cases$mu_K[i]
vif <- test_cases$vif[i]
var_K <- vif * (mu_K - 1)
# A1 mapping
fit <- DPprior_a1(J = J, mu_K = mu_K, var_K = var_K)
# Exact moments with A1 parameters
exact <- exact_K_moments(J, fit$a, fit$b)
data.frame(
J = J,
mu_K_target = mu_K,
var_K_target = var_K,
mu_K_achieved = exact$mean,
var_K_achieved = exact$var,
mu_error_pct = 100 * (exact$mean - mu_K) / mu_K,
var_error_pct = 100 * (exact$var - var_K) / var_K
)
}))
knitr::kable(
results,
digits = c(0, 1, 1, 2, 2, 1, 1),
col.names = c("J", "μ_K (target)", "σ²_K (target)",
"μ_K (achieved)", "σ²_K (achieved)",
"Mean Error %", "Var Error %"),
caption = "A1 approximation error: target vs. achieved moments"
)| J | μ_K (target) | σ²_K (target) | μ_K (achieved) | σ²_K (achieved) | Mean Error % | Var Error % |
|---|---|---|---|---|---|---|
| 25 | 5 | 6.0 | 4.27 | 3.22 | -14.6 | -46.3 |
| 50 | 5 | 6.0 | 4.51 | 3.89 | -9.8 | -35.2 |
| 100 | 5 | 6.0 | 4.67 | 4.37 | -6.6 | -27.2 |
| 200 | 5 | 6.0 | 4.77 | 4.72 | -4.5 | -21.3 |
| 25 | 10 | 13.5 | 6.84 | 4.58 | -31.6 | -66.0 |
| 50 | 10 | 13.5 | 7.64 | 6.21 | -23.6 | -54.0 |
| 100 | 10 | 13.5 | 8.21 | 7.53 | -17.9 | -44.2 |
| 200 | 10 | 13.5 | 8.61 | 8.57 | -13.9 | -36.5 |
| 25 | 5 | 8.0 | 4.21 | 3.86 | -15.8 | -51.7 |
| 50 | 5 | 8.0 | 4.46 | 4.78 | -10.8 | -40.2 |
| 100 | 5 | 8.0 | 4.63 | 5.48 | -7.5 | -31.5 |
| 200 | 5 | 8.0 | 4.74 | 6.00 | -5.2 | -25.0 |
| 25 | 10 | 18.0 | 6.78 | 5.31 | -32.2 | -70.5 |
| 50 | 10 | 18.0 | 7.59 | 7.43 | -24.1 | -58.7 |
| 100 | 10 | 18.0 | 8.16 | 9.24 | -18.4 | -48.7 |
| 200 | 10 | 18.0 | 8.57 | 10.70 | -14.3 | -40.6 |
| 25 | 5 | 12.0 | 4.10 | 4.98 | -17.9 | -58.5 |
| 50 | 5 | 12.0 | 4.37 | 6.40 | -12.6 | -46.7 |
| 100 | 5 | 12.0 | 4.55 | 7.52 | -9.0 | -37.4 |
| 200 | 5 | 12.0 | 4.67 | 8.38 | -6.6 | -30.2 |
| 25 | 10 | 27.0 | 6.67 | 6.67 | -33.3 | -75.3 |
| 50 | 10 | 27.0 | 7.48 | 9.76 | -25.2 | -63.8 |
| 100 | 10 | 27.0 | 8.07 | 12.51 | -19.3 | -53.7 |
| 200 | 10 | 27.0 | 8.49 | 14.79 | -15.1 | -45.2 |
5.3 Systematic Patterns
# Create heatmap of A1 mean errors
J_grid <- c(20, 30, 50, 75, 100, 150, 200)
mu_grid <- seq(3, 15, by = 2)
error_matrix <- matrix(NA, nrow = length(J_grid), ncol = length(mu_grid))
rownames(error_matrix) <- paste0("J=", J_grid)
colnames(error_matrix) <- paste0("μ=", mu_grid)
for (i in seq_along(J_grid)) {
for (j in seq_along(mu_grid)) {
J <- J_grid[i]
mu_K <- mu_grid[j]
if (mu_K < J) {
var_K <- 2 * (mu_K - 1) # VIF = 2
fit <- DPprior_a1(J = J, mu_K = mu_K, var_K = var_K)
exact <- exact_K_moments(J, fit$a, fit$b)
error_matrix[i, j] <- 100 * (exact$mean - mu_K) / mu_K
}
}
}
# Convert to data frame for ggplot
error_df <- expand.grid(J = J_grid, mu_K = mu_grid)
error_df$error <- as.vector(error_matrix)
ggplot(error_df[!is.na(error_df$error), ],
aes(x = factor(mu_K), y = factor(J), fill = error)) +
geom_tile() +
geom_text(aes(label = sprintf("%.1f%%", error)), color = "white", size = 3) +
scale_fill_gradient2(low = "#377EB8", mid = "white", high = "#E41A1C",
midpoint = 0, name = "Mean\nError %") +
labs(
x = expression("Target " * mu[K]),
y = "Sample Size J",
title = "A1 Mean Error Pattern",
subtitle = "VIF = 2 for all cases"
) +
theme_minimal()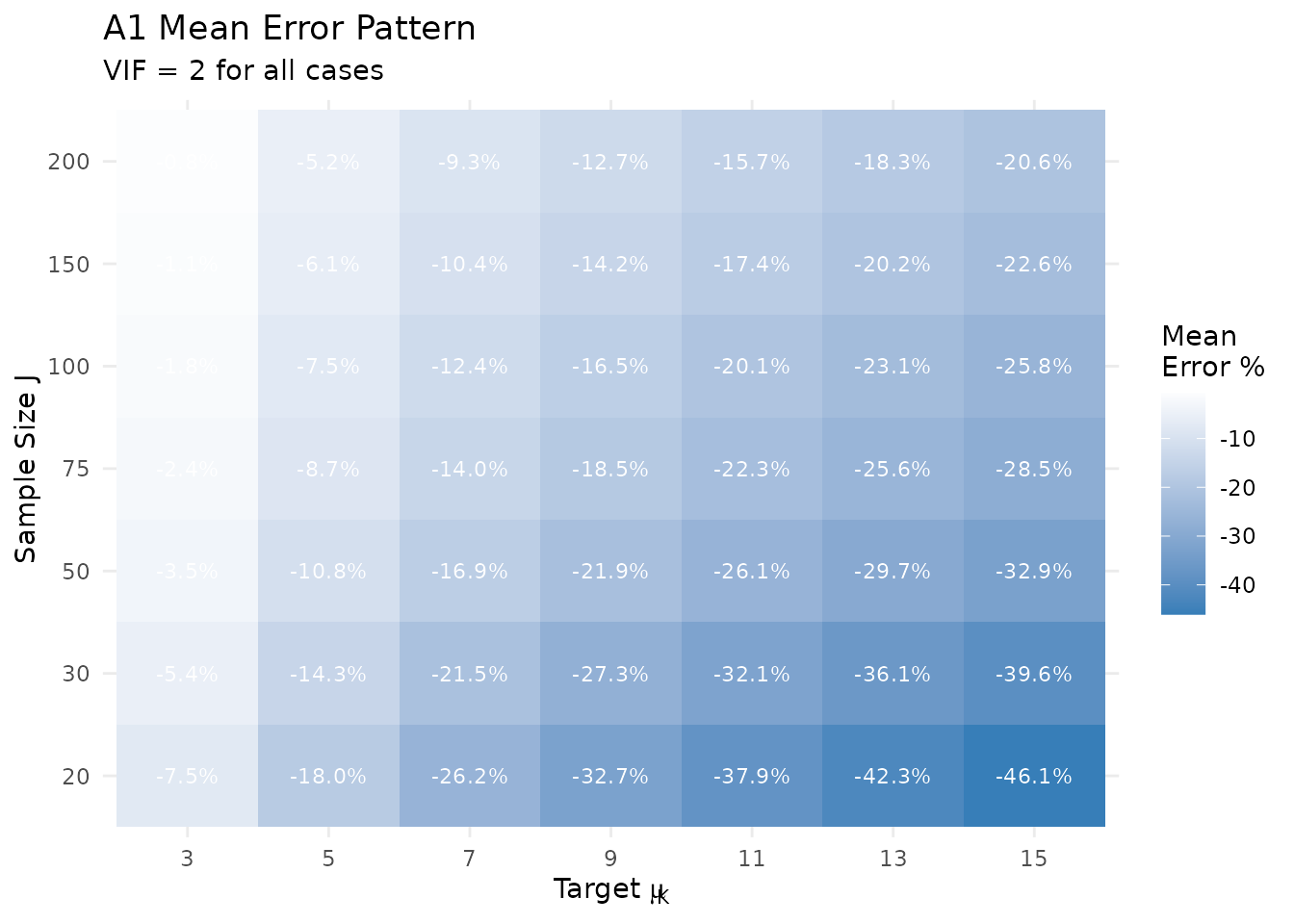
A1 mean error as a function of J and μ_K.
5.4 Critical Warning: NegBin Marginal Limitations
Novel Finding. Tables from the research notes demonstrate that the NegBin marginal approximation can produce:
- Mean errors of 40-75%
- Variance errors of 230-730%
This confirms that A1 should be viewed primarily as an
initializer, not as a final solution. The A2 Newton refinement
(see vignette("theory-newton")) should be applied for any
application requiring accurate moment matching.
# Compare A1 to A2 refinement
J <- 50
mu_K <- 5
var_K <- 8
# A1 only
fit_a1 <- DPprior_a1(J = J, mu_K = mu_K, var_K = var_K)
exact_a1 <- exact_K_moments(J, fit_a1$a, fit_a1$b)
# A2 refinement (full DPprior_fit)
fit_a2 <- DPprior_fit(J = J, mu_K = mu_K, var_K = var_K)
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 48.1% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
exact_a2 <- exact_K_moments(J, fit_a2$a, fit_a2$b)
cat("Comparison: A1 vs A2 Refinement\n")
#> Comparison: A1 vs A2 Refinement
cat(paste(rep("=", 50), collapse = ""), "\n\n")
#> ==================================================
cat(sprintf("Target: μ_K = %.1f, σ²_K = %.1f\n\n", mu_K, var_K))
#> Target: μ_K = 5.0, σ²_K = 8.0
cat("A1 (closed-form):\n")
#> A1 (closed-form):
cat(sprintf(" Parameters: a = %.4f, b = %.4f\n", fit_a1$a, fit_a1$b))
#> Parameters: a = 4.0000, b = 3.9120
cat(sprintf(" Achieved: μ_K = %.4f, σ²_K = %.4f\n", exact_a1$mean, exact_a1$var))
#> Achieved: μ_K = 4.4614, σ²_K = 4.7831
cat(sprintf(" Errors: %.2f%% (mean), %.2f%% (var)\n\n",
100 * (exact_a1$mean - mu_K) / mu_K,
100 * (exact_a1$var - var_K) / var_K))
#> Errors: -10.77% (mean), -40.21% (var)
cat("A2 (Newton refinement):\n")
#> A2 (Newton refinement):
cat(sprintf(" Parameters: a = %.4f, b = %.4f\n", fit_a2$a, fit_a2$b))
#> Parameters: a = 2.0361, b = 1.6051
cat(sprintf(" Achieved: μ_K = %.6f, σ²_K = %.6f\n", exact_a2$mean, exact_a2$var))
#> Achieved: μ_K = 5.000000, σ²_K = 8.000000
cat(sprintf(" Errors: %.2e%% (mean), %.2e%% (var)\n",
100 * abs(exact_a2$mean - mu_K) / mu_K,
100 * abs(exact_a2$var - var_K) / var_K))
#> Errors: 1.66e-08% (mean), 9.44e-08% (var)6. Summary
This vignette has provided a rigorous treatment of the A1 approximation theory:
Poisson approximation: The classical result that converges to Poisson provides the theoretical foundation.
Scaling constant: The default provides the best mean-level accuracy for typical values.
NegBin mixture: When , the marginal approximately follows a Negative Binomial distribution.
Inverse mapping: Theorem 1 provides closed-form expressions for given target moments .
Limitations: The A1 approximation can have substantial errors and should be viewed as an initializer for the A2 Newton refinement.
References
Abramowitz, M., & Stegun, I. A. (1964). Handbook of Mathematical Functions. National Bureau of Standards.
Antoniak, C. E. (1974). Mixtures of Dirichlet processes with applications to Bayesian nonparametric problems. The Annals of Statistics, 2(6), 1152-1174.
Arratia, R., Barbour, A. D., & Tavaré, S. (2000). The number of components in a logarithmic combinatorial structure. The Annals of Applied Probability, 10(2), 331-361.
Dorazio, R. M. (2009). On selecting a prior for the precision parameter of Dirichlet process mixture models. Journal of Statistical Planning and Inference, 139(10), 3384-3390.
Murugiah, S., & Sweeting, T. J. (2012). Selecting the precision parameter prior in Dirichlet process mixture models. Journal of Statistical Computation and Simulation, 82(9), 1307-1322.
Vicentini, C., & Jermyn, I. H. (2025). Prior selection for the precision parameter of Dirichlet process mixtures. arXiv:2502.00864.
For questions about this vignette or the DPprior package, please visit the GitHub repository.