Quick Start: Your First Prior in 5 Minutes
JoonHo Lee
2026-01-01
Source:vignettes/quick-start.Rmd
quick-start.RmdOverview
This vignette demonstrates the fastest path to eliciting a Gamma hyperprior for the concentration parameter in a Dirichlet Process mixture model. By the end of this 5-minute tutorial, you will be able to:
- Specify your prior belief about the number of clusters
- Convert that belief into Gamma hyperparameters
- Visualize and verify your elicited prior
1. Minimal Example
Let’s walk through a concrete scenario. Suppose you are analyzing a multisite educational trial with 50 sites, and you expect that these sites will cluster into approximately 5 distinct effect groups. This setup mirrors the “DP-inform” prior elicitation approach used in Lee et al. (2025), who demonstrated the benefits of informative Dirichlet Process priors for estimating site-specific effects in multisite trials. While that paper relied on chi-square discrepancy measures with computationally intensive grid search, the DPprior package provides instant, closed-form solutions for the same elicitation task.
library(DPprior)
# Just two inputs needed:
# 1. J: Number of sites (or observations for clustering)
# 2. mu_K: Expected number of clusters
fit <- DPprior_fit(J = 50, mu_K = 5, confidence = "medium")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 49.7% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
# View the result
print(fit)
#> DPprior Prior Elicitation Result
#> =============================================
#>
#> Gamma Hyperprior: α ~ Gamma(a = 1.4082, b = 1.0770)
#> E[α] = 1.308, SD[α] = 1.102
#>
#> Target (J = 50):
#> E[K_J] = 5.00
#> Var(K_J) = 10.00
#> (from confidence = 'medium')
#>
#> Achieved:
#> E[K_J] = 5.000000, Var(K_J) = 10.000000
#> Residual = 3.94e-10
#>
#> Method: A2-MN (7 iterations)
#>
#> Dominance Risk: HIGH ✘ (P(w₁>0.5) = 50%)That’s it! You now have a principled Gamma hyperprior for .
2. Understanding the Output
The DPprior_fit() function returns an object containing
the Gamma hyperparameters and diagnostic information. Let’s examine the
key components:
# The Gamma hyperparameters
cat("Gamma shape (a):", round(fit$a, 4), "\n")
#> Gamma shape (a): 1.4082
cat("Gamma rate (b):", round(fit$b, 4), "\n")
#> Gamma rate (b): 1.077
# What these imply about alpha
alpha_mean <- fit$a / fit$b
alpha_sd <- sqrt(fit$a) / fit$b
cat("\nPrior mean of α:", round(alpha_mean, 3), "\n")
#>
#> Prior mean of α: 1.308
cat("Prior SD of α: ", round(alpha_sd, 3), "\n")
#> Prior SD of α: 1.102
# The target specification
cat("\nTarget E[K]:", fit$target$mu_K, "\n")
#>
#> Target E[K]: 5
cat("Target Var(K):", round(fit$target$var_K, 2), "\n")
#> Target Var(K): 10Interpretation: The elicited prior implies that you expect around 5 clusters when sampling observations, with uncertainty captured by the variance.
3. Visualizing Your Prior
The DPprior package provides built-in visualization functions to help you understand and communicate your elicited prior.
3.1 Full Dashboard View
The plot() method displays a comprehensive four-panel
dashboard:
plot(fit)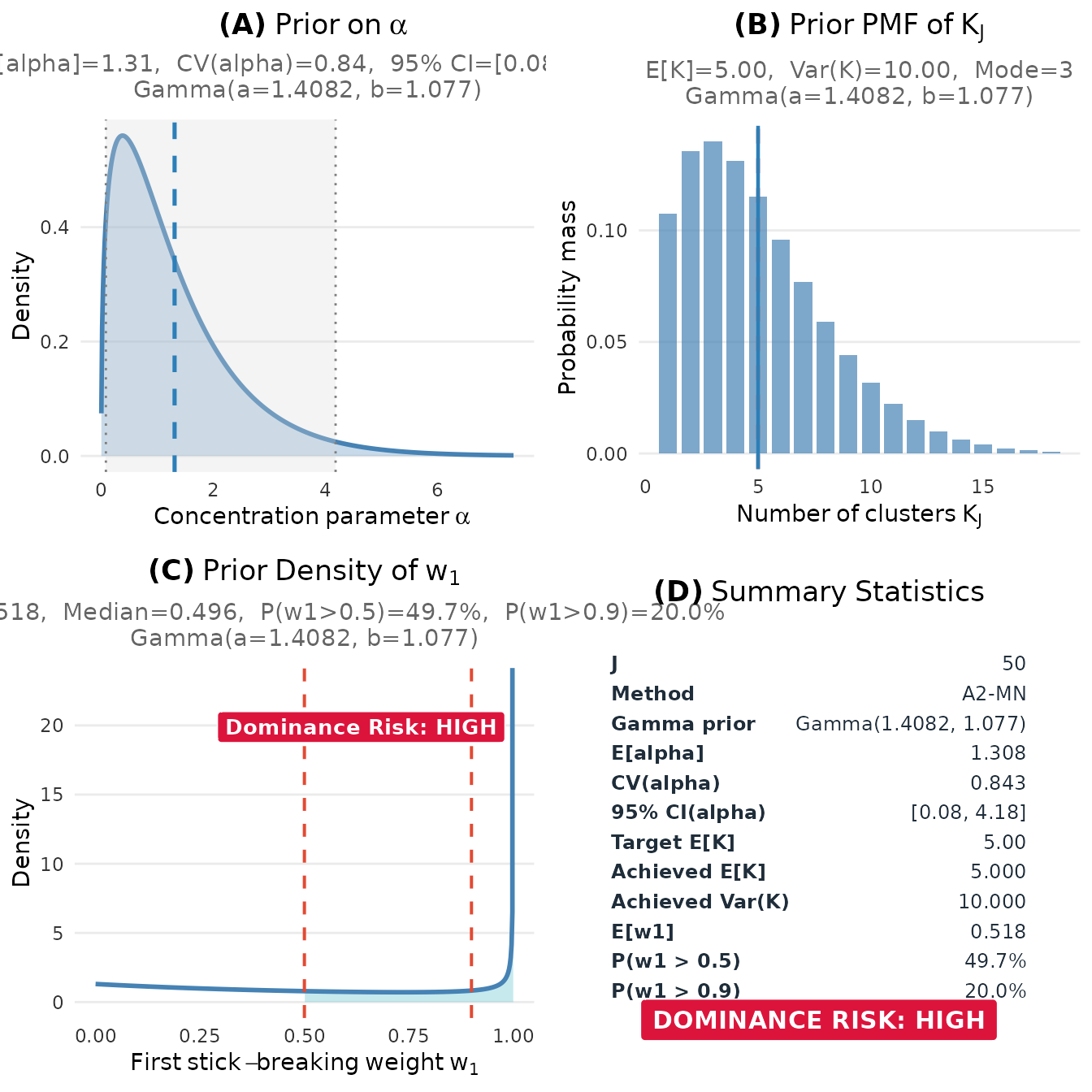
Prior elicitation dashboard showing the distributions of α, K, and w₁.
#> TableGrob (2 x 2) "dpprior_dashboard": 4 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) dpprior_dashboard gtable[layout]
#> 2 2 (2-2,1-1) dpprior_dashboard gtable[layout]
#> 3 3 (1-1,2-2) dpprior_dashboard gtable[layout]
#> 4 4 (2-2,2-2) dpprior_dashboard gtable[layout]The dashboard includes:
- Panel (A): The Gamma prior density for the concentration parameter , with key statistics (mean, CV, 95% CI)
- Panel (B): The implied marginal PMF of the number of clusters , showing E[K], Var(K), and mode
- Panel (C): The prior density of the first stick-breaking weight , with dominance risk assessment (useful for detecting overly concentrated priors)
- Panel (D): Summary statistics table consolidating all key diagnostics
3.2 Individual Plots
You can also create individual plots for more focused analysis:
plot_alpha_prior(fit)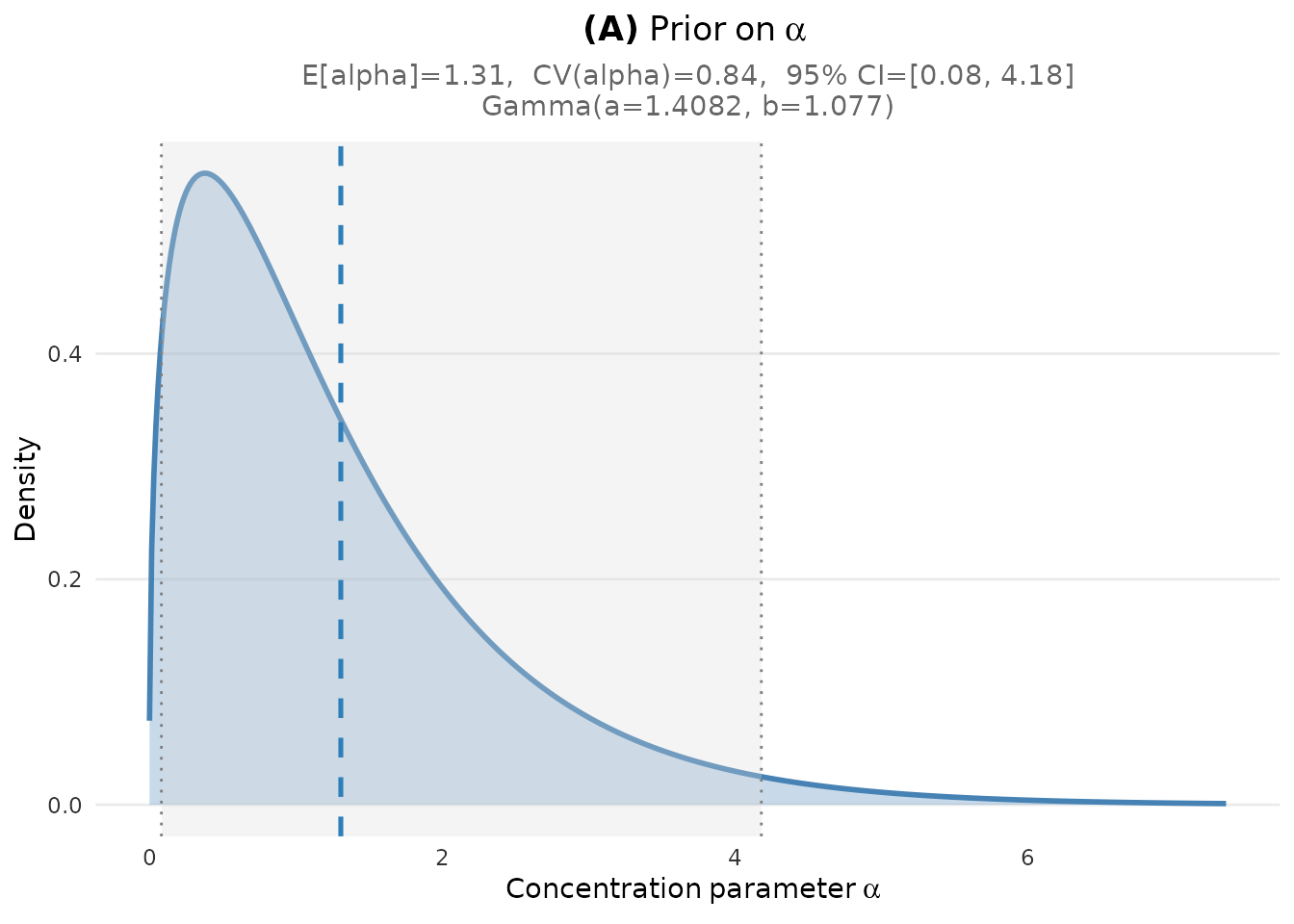
plot_K_prior(fit)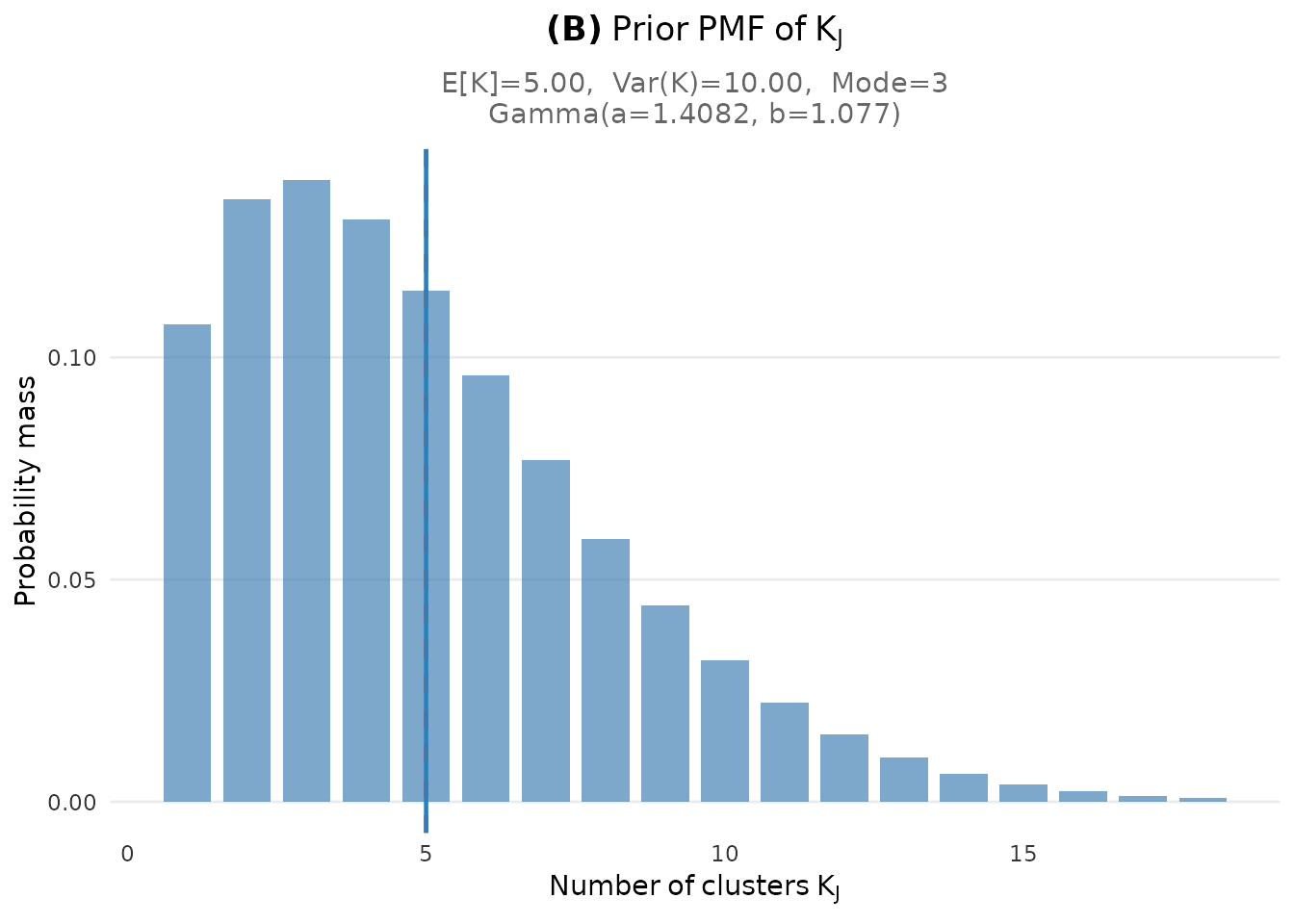
plot_w1_prior(fit)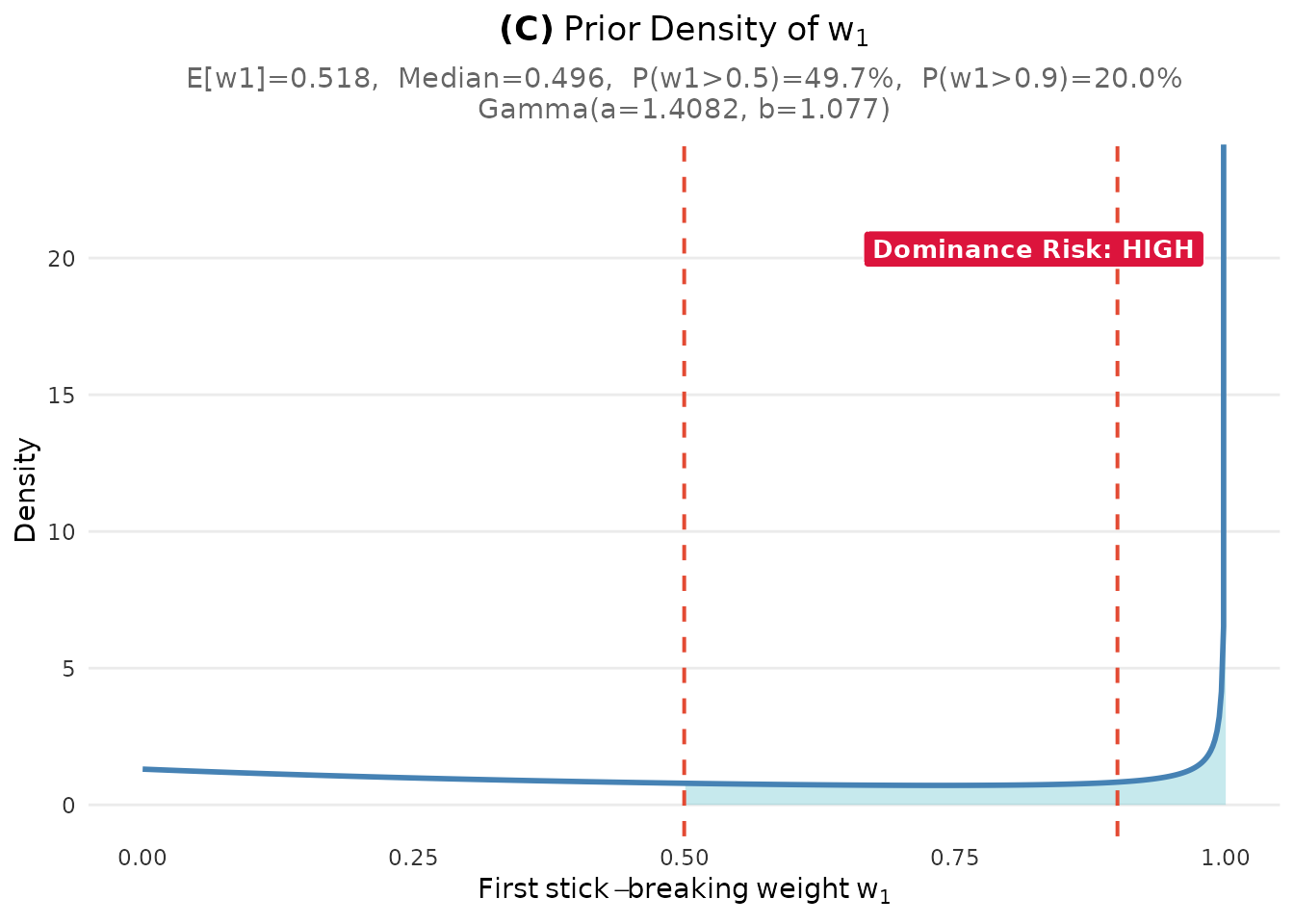
4. Three Ways to Specify Your Uncertainty
The DPprior_fit() function offers flexibility in how you
express your prior uncertainty about the number of clusters:
Method 1: Confidence Level (Recommended for Beginners)
The simplest approach uses qualitative confidence levels:
# Low confidence = high uncertainty (wide prior)
fit_low <- DPprior_fit(J = 50, mu_K = 5, confidence = "low")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 56.3% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
# Medium confidence = moderate uncertainty (default)
fit_med <- DPprior_fit(J = 50, mu_K = 5, confidence = "medium")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 49.7% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
# High confidence = low uncertainty (concentrated prior)
fit_high <- DPprior_fit(J = 50, mu_K = 5, confidence = "high")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 46.5% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
# Compare the resulting Gamma parameters
comparison <- data.frame(
Confidence = c("Low", "Medium", "High"),
var_K = round(c(fit_low$target$var_K, fit_med$target$var_K,
fit_high$target$var_K), 2),
a = round(c(fit_low$a, fit_med$a, fit_high$a), 3),
b = round(c(fit_low$b, fit_med$b, fit_high$b), 3),
CV_alpha = round(1/sqrt(c(fit_low$a, fit_med$a, fit_high$a)), 3)
)
comparison
#> Confidence var_K a b CV_alpha
#> 1 Low 20 0.518 0.341 1.390
#> 2 Medium 10 1.408 1.077 0.843
#> 3 High 6 3.568 2.900 0.529Interpretation: Higher confidence (lower uncertainty) leads to a more concentrated prior on , reflected in the lower coefficient of variation (CV).
Method 2: Direct Variance Specification
For users who want precise control:
# Specify variance of K directly
fit_direct <- DPprior_fit(J = 50, mu_K = 5, var_K = 10)
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 49.7% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
cat("Direct specification: var_K = 10\n")
#> Direct specification: var_K = 10
cat(" Gamma(a =", round(fit_direct$a, 3), ", b =", round(fit_direct$b, 3), ")\n")
#> Gamma(a = 1.408 , b = 1.077 )Method 3: Different Calibration Methods
For advanced users, different calibration algorithms are available:
# A2-MN (default): Exact moment matching via Newton's method
fit_newton <- DPprior_fit(J = 50, mu_K = 5, var_K = 10, method = "A2-MN")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 49.7% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
# A1: Fast closed-form approximation (good for large J)
fit_approx <- DPprior_fit(J = 50, mu_K = 5, var_K = 10, method = "A1")
#> Warning: HIGH DOMINANCE RISK: P(w1 > 0.5) = 53.3% exceeds 40%.
#> This may indicate unintended prior behavior (RN-07).
#> Consider using DPprior_dual() for weight-constrained elicitation.
#> See ?DPprior_diagnostics for interpretation.
cat("A2-MN: Gamma(", round(fit_newton$a, 3), ", ", round(fit_newton$b, 3), ")\n", sep = "")
#> A2-MN: Gamma(1.408, 1.077)
cat("A1: Gamma(", round(fit_approx$a, 3), ", ", round(fit_approx$b, 3), ")\n", sep = "")
#> A1: Gamma(2.667, 2.608)5. Using Your Prior in Practice
Once you have elicited your Gamma hyperparameters, you can use them in your Bayesian software of choice.
R (for simulation)
# Draw samples from the elicited prior
n_samples <- 10000
alpha_samples <- rgamma(n_samples, shape = fit$a, rate = fit$b)
cat("Summary of sampled α values:\n")
#> Summary of sampled α values:
cat(" Mean:", round(mean(alpha_samples), 3), "\n")
#> Mean: 1.32
cat(" SD: ", round(sd(alpha_samples), 3), "\n")
#> SD: 1.118
cat(" 95% CI: [", round(quantile(alpha_samples, 0.025), 3), ", ",
round(quantile(alpha_samples, 0.975), 3), "]\n", sep = "")
#> 95% CI: [0.079, 4.256]What’s Next?
This quick start covered the essentials. For more advanced topics:
- Applied Guide: Comprehensive elicitation workflow with sensitivity analysis
- Dual-Anchor Framework: Control both cluster count AND cluster weight concentration
- Diagnostics: Verify your prior meets your specifications
- Case Studies: Real-world examples from education, medicine, and policy research
Summary
| Step | Action | Code |
|---|---|---|
| 1 | Define context | J <- 50 |
| 2 | Set expected clusters | mu_K <- 5 |
| 3 | Choose confidence | confidence = "medium" |
| 4 | Elicit prior | fit <- DPprior_fit(J, mu_K, confidence) |
| 5 | Visualize | plot(fit) |
| 6 | Use in model | alpha ~ gamma(fit$a, fit$b) |
References
Lee, J., Che, J., Rabe-Hesketh, S., Feller, A., & Miratrix, L. (2025). Improving the estimation of site-specific effects and their distribution in multisite trials. Journal of Educational and Behavioral Statistics, 50(5), 731–764. https://doi.org/10.3102/10769986241254286
For questions or feedback, please visit the GitHub repository.