Overview
The sim_item_params() function generates item parameters
(difficulty
and discrimination
)
for IRT simulation studies. It is designed with four key principles:
- Realistic difficulties: Integration with the Item Response Warehouse (IRW) for empirically-grounded difficulty distributions
- Correlated parameters: Support for the empirically-observed negative correlation between difficulty and discrimination
- Marginal preservation: The copula method preserves exact marginal distributions
- Reliability targeting: A scale factor enables subsequent calibration for target reliability
This vignette covers:
- Sources for difficulty generation
- Methods for discrimination generation
- The difficulty-discrimination correlation
- Generating multiple test forms
- Integration with reliability-targeted calibration
The Difficulty-Discrimination Correlation
A critical finding from psychometric research is that item difficulty and discrimination are negatively correlated in real assessments (Sweeney et al., 2022). That is:
- Easy items ( low) tend to have higher discrimination ( high)
- Difficult items ( high) tend to have lower discrimination ( low)
This correlation, typically around , has important implications:
- Ignoring it produces unrealistic simulation data
- Standard independent generation misses this structural feature
- The correlation affects test information functions
sim_item_params() handles this by default using the
copula method with rho = -0.3.
Basic Usage
Rasch Model
For the Rasch model, all discriminations are set to 1:
# Generate 25 Rasch items with IRW difficulties
items_rasch <- sim_item_params(
n_items = 25,
model = "rasch",
source = "irw",
seed = 42
)
print(items_rasch)
#> Item Parameters Object
#> ======================
#> Model : RASCH
#> Source : irw
#> Items per form : 25
#> Number of forms: 1
#> Scale factor : 1.000
#> Centered : Yes
#>
#> Difficulty (beta):
#> Mean: -0.0000, SD: 0.7992, Range: [-1.484, 1.469]2PL Model
For the 2PL model, both difficulties and discriminations are generated:
# Generate 30 2PL items with correlated parameters
items_2pl <- sim_item_params(
n_items = 30,
model = "2pl",
source = "irw",
method = "copula",
seed = 42
)
print(items_2pl)
#> Item Parameters Object
#> ======================
#> Model : 2PL
#> Source : irw
#> Method : copula
#> Items per form : 30
#> Number of forms: 1
#> Scale factor : 1.000
#> Centered : Yes
#>
#> Difficulty (beta):
#> Mean: -0.0000, SD: 0.8357, Range: [-1.607, 1.463]
#>
#> Discrimination (lambda, scaled):
#> Mean: 1.0865, SD: 0.4031, Range: [0.590, 2.107]
#>
#> Correlation (beta, log-lambda):
#> Target (rho): -0.300
#> Achieved Pearson : -0.330
#> Achieved Spearman: -0.466Notice the achieved correlation between and is close to the target of -0.3.
Sources for Difficulty Generation
sim_item_params() supports four sources for generating
item difficulties:
1. IRW (Item Response Warehouse)
The recommended source for realistic simulations. IRW provides empirically-grounded difficulty distributions based on thousands of real assessment items.
items_irw <- sim_item_params(
n_items = 25,
model = "rasch",
source = "irw",
seed = 42
)
# Examine difficulty distribution
summary(items_irw$data$beta)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> -1.48424 -0.54581 -0.08185 0.00000 0.48541 1.46931Requirements: The irw package must be
installed:
# Install IRW package from GitHub
devtools::install_github("itemresponsewarehouse/irw")2. Parametric
Generate difficulties from a parametric distribution:
# Normal distribution
items_normal <- sim_item_params(
n_items = 25,
model = "rasch",
source = "parametric",
difficulty_params = list(
mu = 0,
sigma = 1,
distribution = "normal"
),
seed = 42
)
# Uniform distribution
items_uniform <- sim_item_params(
n_items = 25,
model = "rasch",
source = "parametric",
difficulty_params = list(
mu = 0,
sigma = 1,
distribution = "uniform"
),
seed = 42
)3. Hierarchical
Joint bivariate normal generation following Glas & van der Linden (2003). Both and are drawn from a multivariate normal:
items_hier <- sim_item_params(
n_items = 25,
model = "2pl",
source = "hierarchical",
hierarchical_params = list(
mu = c(0, 0), # Means: (log-lambda, beta)
tau = c(0.25, 1), # SDs
rho = -0.3 # Correlation
),
seed = 42
)
print(items_hier)
#> Item Parameters Object
#> ======================
#> Model : 2PL
#> Source : hierarchical
#> Items per form : 25
#> Number of forms: 1
#> Scale factor : 1.000
#> Centered : Yes
#>
#> Difficulty (beta):
#> Mean: -0.0000, SD: 1.3077, Range: [-2.863, 2.109]
#>
#> Discrimination (lambda, scaled):
#> Mean: 1.0776, SD: 0.2710, Range: [0.720, 1.812]
#>
#> Correlation (beta, log-lambda):
#> Target (rho): -0.300
#> Achieved Pearson : -0.345
#> Achieved Spearman: -0.3754. Custom
Supply your own parameters directly:
# Custom difficulties and discriminations
items_custom <- sim_item_params(
n_items = 10,
model = "2pl",
source = "custom",
custom_params = list(
beta = seq(-2, 2, length.out = 10),
lambda = rep(1.2, 10)
),
seed = 42
)
#> Warning in cor(df$beta, log(df$lambda_unscaled)): the standard deviation is
#> zero
#> Warning in cor(df$beta, log(df$lambda_unscaled), method = "spearman"): the
#> standard deviation is zero
#> Warning in cor(data$beta, log(data$lambda_unscaled)): the standard deviation is
#> zero
#> Warning in cor(data$beta, log(data$lambda_unscaled), method = "spearman"): the
#> standard deviation is zero
items_custom$data
#> form_id item_id beta lambda lambda_unscaled
#> 1 1 1 -2.0000000 1.2 1.2
#> 2 1 2 -1.5555556 1.2 1.2
#> 3 1 3 -1.1111111 1.2 1.2
#> 4 1 4 -0.6666667 1.2 1.2
#> 5 1 5 -0.2222222 1.2 1.2
#> 6 1 6 0.2222222 1.2 1.2
#> 7 1 7 0.6666667 1.2 1.2
#> 8 1 8 1.1111111 1.2 1.2
#> 9 1 9 1.5555556 1.2 1.2
#> 10 1 10 2.0000000 1.2 1.2You can also provide functions that generate parameters:
items_custom_fn <- sim_item_params(
n_items = 20,
model = "2pl",
source = "custom",
custom_params = list(
beta = function(n) rnorm(n, 0, 1.5),
lambda = function(n) rlnorm(n, 0, 0.3)
),
seed = 42
)Methods for Discrimination Generation
When using source = "irw" or
source = "parametric" with model = "2pl", you
need to specify how discriminations are generated. Three methods are
available:
1. Copula Method (Recommended)
The Gaussian copula method preserves exact marginal distributions while achieving the target correlation:
Algorithm:
- Transform to uniform via empirical CDF:
- Transform to normal:
- Generate correlated normal:
- Transform to uniform:
- Transform to log-normal:
items_copula <- sim_item_params(
n_items = 100,
model = "2pl",
source = "irw",
method = "copula",
discrimination_params = list(
mu_log = 0, # Mean of log(lambda)
sigma_log = 0.3, # SD of log(lambda)
rho = -0.3 # Target correlation
),
seed = 42
)
# Check achieved correlation
cat(sprintf("Target rho: -0.30\n"))
#> Target rho: -0.30
cat(sprintf("Achieved Spearman: %.3f\n",
items_copula$achieved$overall$cor_spearman_pooled))
#> Achieved Spearman: -0.191Why Copula is Recommended:
- Preserves the exact IRW difficulty distribution
- Guarantees log-normal marginal for discriminations
- Achieves target Spearman correlation (robust to non-normality)
- Works well with any difficulty distribution shape
2. Conditional Method
Uses conditional normal regression:
items_cond <- sim_item_params(
n_items = 100,
model = "2pl",
source = "irw",
method = "conditional",
discrimination_params = list(rho = -0.3),
seed = 42
)
cat(sprintf("Achieved Pearson: %.3f\n",
items_cond$achieved$overall$cor_pearson_pooled))
#> Achieved Pearson: -0.248
cat(sprintf("Achieved Spearman: %.3f\n",
items_cond$achieved$overall$cor_spearman_pooled))
#> Achieved Spearman: -0.207Note: The conditional method assumes linear relationships and normal errors. When IRW difficulties are non-normal, achieved correlations may differ from targets.
3. Independent Method
Generates discriminations independently of difficulties (no correlation):
items_indep <- sim_item_params(
n_items = 100,
model = "2pl",
source = "irw",
method = "independent",
seed = 42
)
cat(sprintf("Achieved correlation: %.3f (expected: ~0)\n",
items_indep$achieved$overall$cor_spearman_pooled))
#> Achieved correlation: 0.051 (expected: ~0)Customizing Discrimination Parameters
The discrimination_params list controls the log-normal
distribution of discriminations:
# Higher average discrimination
items_high_disc <- sim_item_params(
n_items = 30,
model = "2pl",
source = "irw",
method = "copula",
discrimination_params = list(
mu_log = 0.3, # E[lambda] ≈ exp(0.3) ≈ 1.35
sigma_log = 0.25, # Less variation
rho = -0.3
),
seed = 42
)
cat(sprintf("Mean lambda: %.3f\n", mean(items_high_disc$data$lambda)))
#> Mean lambda: 1.435
cat(sprintf("SD lambda: %.3f\n", sd(items_high_disc$data$lambda)))
#> SD lambda: 0.435Generating Multiple Test Forms
Generate multiple parallel forms with independent item samples:
items_5forms <- sim_item_params(
n_items = 20,
model = "2pl",
source = "irw",
method = "copula",
n_forms = 5,
seed = 42
)
# Check structure
cat(sprintf("Total items: %d\n", nrow(items_5forms$data)))
#> Total items: 100
cat(sprintf("Items per form: %d\n", items_5forms$n_items))
#> Items per form: 20
cat(sprintf("Number of forms: %d\n", items_5forms$n_forms))
#> Number of forms: 5
# View first few rows
head(items_5forms$data, 10)
#> form_id item_id beta lambda lambda_unscaled
#> 1 1 1 -0.31979479 0.9134159 0.9134159
#> 2 1 2 -0.13448739 1.1691256 1.1691256
#> 3 1 3 1.21101694 0.6461936 0.6461936
#> 4 1 4 0.07285940 1.2932730 1.2932730
#> 5 1 5 -1.41375360 1.2920870 1.2920870
#> 6 1 6 0.49929409 0.9477748 0.9477748
#> 7 1 7 -0.86741035 1.0869113 1.0869113
#> 8 1 8 0.71169376 0.9945658 0.9945658
#> 9 1 9 0.58379081 0.7129531 0.7129531
#> 10 1 10 -0.04531184 1.0322409 1.0322409Per-Form Statistics
# Access per-form statistics
for (f in 1:3) {
stats <- items_5forms$achieved$by_form[[f]]
cat(sprintf("Form %d: beta_mean=%.3f, lambda_mean=%.3f, cor=%.3f\n",
f, stats$beta_mean, stats$lambda_mean, stats$cor_spearman))
}
#> Form 1: beta_mean=0.000, lambda_mean=1.027, cor=-0.540
#> Form 2: beta_mean=0.000, lambda_mean=1.152, cor=-0.202
#> Form 3: beta_mean=0.000, lambda_mean=1.109, cor=-0.320The Scale Parameter
The scale parameter is crucial for reliability-targeted
simulation. It multiplies all discriminations by a constant factor:
where is the baseline discrimination and is the scale factor.
# Baseline (scale = 1)
items_base <- sim_item_params(
n_items = 25,
model = "2pl",
source = "irw",
scale = 1,
seed = 42
)
# Scaled up (scale = 1.5)
items_scaled <- sim_item_params(
n_items = 25,
model = "2pl",
source = "irw",
scale = 1.5,
seed = 42
)
cat(sprintf("Baseline mean lambda: %.3f\n", mean(items_base$data$lambda)))
#> Baseline mean lambda: 1.026
cat(sprintf("Scaled mean lambda: %.3f\n", mean(items_scaled$data$lambda)))
#> Scaled mean lambda: 1.539
cat(sprintf("Ratio: %.2f\n",
mean(items_scaled$data$lambda) / mean(items_base$data$lambda)))
#> Ratio: 1.50Unscaled Lambda
The output always includes lambda_unscaled for
reference:
# Both are stored
head(items_scaled$data[, c("lambda", "lambda_unscaled")])
#> lambda lambda_unscaled
#> 1 1.556978 1.0379855
#> 2 1.496971 0.9979810
#> 3 1.501281 1.0008540
#> 4 1.152807 0.7685377
#> 5 1.852231 1.2348209
#> 6 1.592726 1.0618176
# Verify relationship
all.equal(
items_scaled$data$lambda,
items_scaled$data$lambda_unscaled * items_scaled$scale
)
#> [1] TRUECentering Difficulties
By default, difficulties are centered to sum to zero (for model identification):
# Default: centered
items_centered <- sim_item_params(
n_items = 25,
model = "rasch",
source = "irw",
center_difficulties = TRUE,
seed = 42
)
# Uncentered
items_uncentered <- sim_item_params(
n_items = 25,
model = "rasch",
source = "irw",
center_difficulties = FALSE,
seed = 42
)
cat(sprintf("Centered mean: %.6f\n", mean(items_centered$data$beta)))
#> Centered mean: -0.000000
cat(sprintf("Uncentered mean: %.6f\n", mean(items_uncentered$data$beta)))
#> Uncentered mean: 0.018108Visualization
The plot() method provides diagnostic
visualizations:
items_viz <- sim_item_params(
n_items = 50,
model = "2pl",
source = "irw",
method = "copula",
seed = 42
)
# Scatter plot with regression line
plot(items_viz, type = "scatter")
#> `geom_smooth()` using formula = 'y ~ x'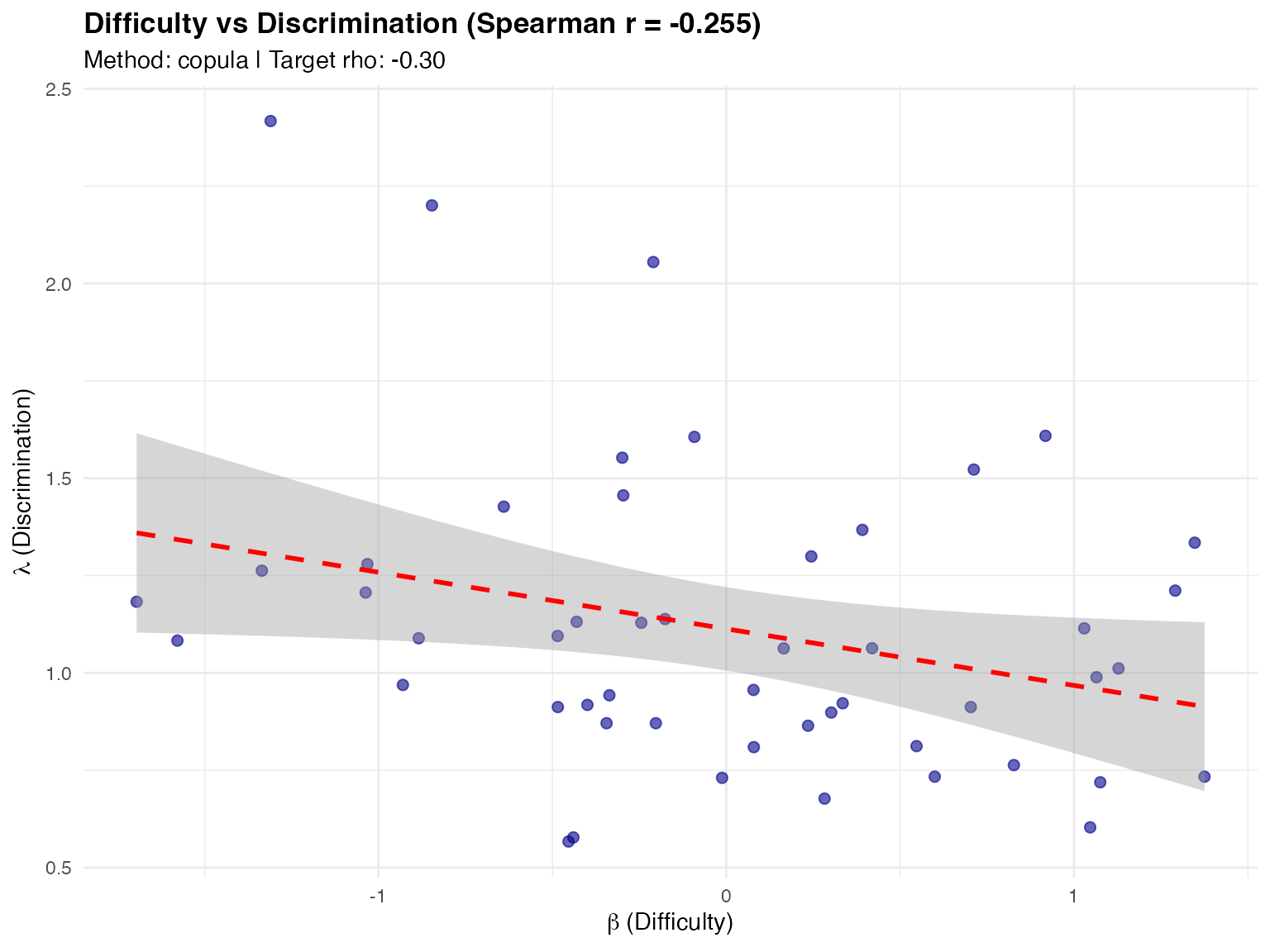
# Density plots
plot(items_viz, type = "density")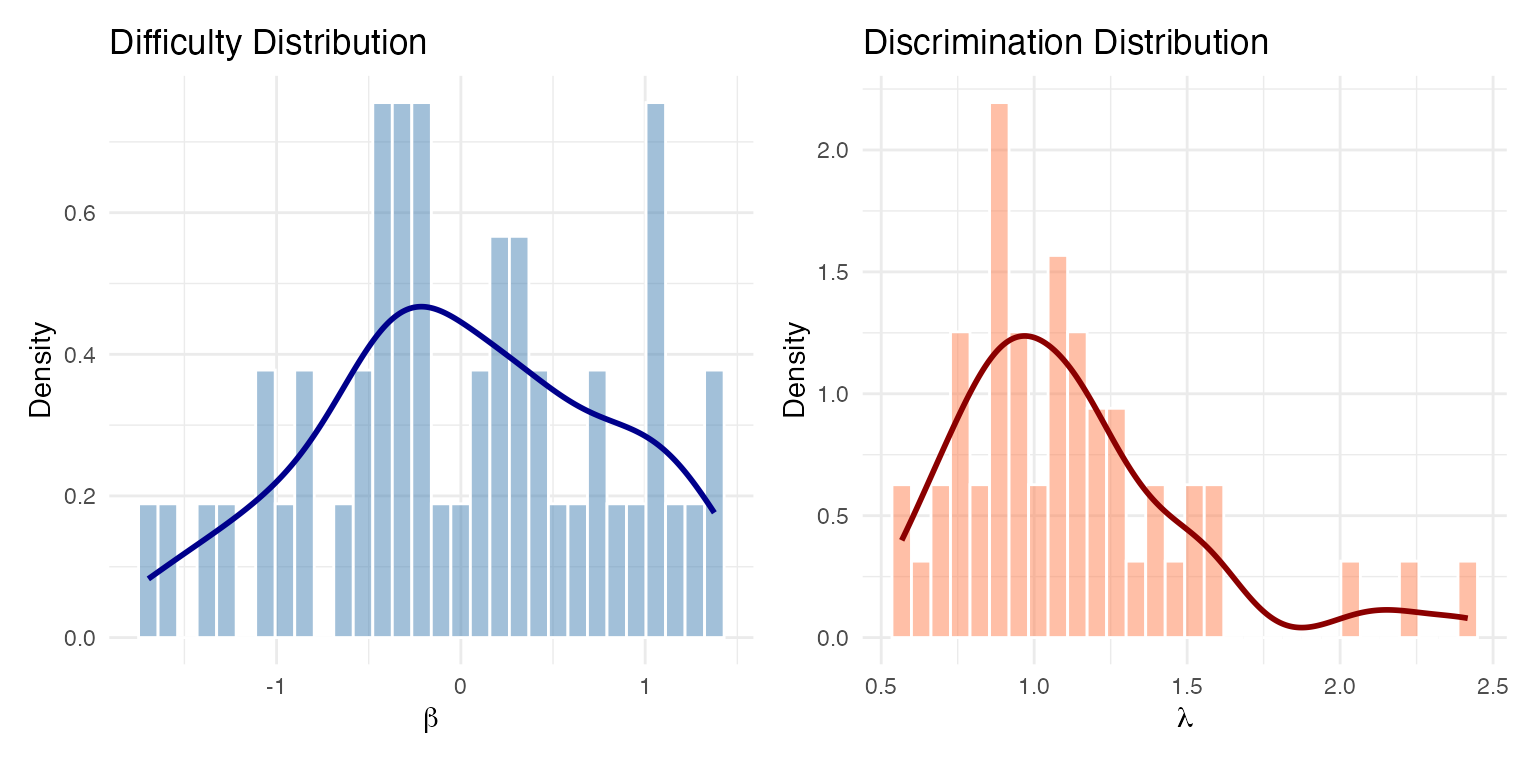
# Combined view (requires patchwork package)
plot(items_viz, type = "both")Integration with eqc_calibrate
In the reliability-targeted simulation framework,
sim_item_params() is called internally by
eqc_calibrate(). You specify item generation settings
through the item_source and item_params
arguments:
# EQC automatically calls sim_item_params internally
eqc_result <- eqc_calibrate(
target_rho = 0.80,
n_items = 25,
model = "2pl",
latent_shape = "normal",
item_source = "irw",
item_params = list(
discrimination_params = list(
mu_log = 0,
sigma_log = 0.3,
rho = -0.3
)
),
seed = 42
)
# The calibrated items are accessible
print(eqc_result$items_calib)Working with the Output Object
The item_params object contains rich information:
items <- sim_item_params(n_items = 25, model = "2pl", source = "irw", seed = 42)
# Structure
names(items)
#> [1] "data" "model" "source" "method" "n_items" "n_forms"
#> [7] "scale" "centered" "params" "achieved"
# Extract as data frame
df <- as.data.frame(items)
head(df)
#> form_id item_id beta lambda lambda_unscaled
#> 1 1 1 -0.390285351 1.0379855 1.0379855
#> 2 1 2 -0.204977951 0.9979810 0.9979810
#> 3 1 3 1.140526381 1.0008540 1.0008540
#> 4 1 4 0.002368838 0.7685377 0.7685377
#> 5 1 5 -1.484244154 1.2348209 1.2348209
#> 6 1 6 0.428803529 1.0618176 1.0618176
# Achieved statistics
items$achieved$overall
#> $n_total
#> [1] 25
#>
#> $beta_mean
#> [1] -2.664535e-17
#>
#> $beta_sd
#> [1] 0.7991656
#>
#> $lambda_mean
#> [1] 1.025717
#>
#> $lambda_sd
#> [1] 0.3127527
#>
#> $cor_pearson_pooled
#> [1] -0.2009709
#>
#> $cor_spearman_pooled
#> [1] -0.3484615Comparison of Methods
Let’s compare the three discrimination generation methods:
set.seed(123)
methods <- c("copula", "conditional", "independent")
results <- list()
for (m in methods) {
results[[m]] <- sim_item_params(
n_items = 200,
model = "2pl",
source = "irw",
method = m,
discrimination_params = list(rho = -0.4),
seed = 123
)
}
# Compare achieved correlations
cat("Method Comparison (target rho = -0.4):\n")
#> Method Comparison (target rho = -0.4):
cat("======================================\n")
#> ======================================
for (m in methods) {
cat(sprintf("%-12s: Pearson = %+.3f, Spearman = %+.3f\n",
m,
results[[m]]$achieved$overall$cor_pearson_pooled,
results[[m]]$achieved$overall$cor_spearman_pooled))
}
#> copula : Pearson = -0.526, Spearman = -0.527
#> conditional : Pearson = -0.495, Spearman = -0.466
#> independent : Pearson = -0.077, Spearman = -0.045The copula method achieves the target Spearman correlation most reliably.
Summary Table: Sources and Methods
| Source | Description | Best For |
|---|---|---|
irw |
Item Response Warehouse | Realistic simulations |
parametric |
Normal/uniform difficulties | Controlled experiments |
hierarchical |
Joint MVN generation | Bayesian framework |
custom |
User-supplied parameters | Specific scenarios |
| Method | Preserves Marginals | Target Correlation | Notes |
|---|---|---|---|
copula |
Yes | Spearman | Recommended |
conditional |
No | Pearson | Assumes normality |
independent |
Yes | None (0) | No correlation |
Practical Recommendations
For Realistic Simulations
items <- sim_item_params(
n_items = 30,
model = "2pl",
source = "irw", # Use IRW
method = "copula", # Preserve marginals
discrimination_params = list(
mu_log = 0,
sigma_log = 0.3,
rho = -0.3 # Empirical correlation
),
seed = 42
)For Controlled Experiments
items <- sim_item_params(
n_items = 25,
model = "2pl",
source = "parametric",
difficulty_params = list(mu = 0, sigma = 1),
method = "conditional",
discrimination_params = list(
mu_log = 0,
sigma_log = 0.25,
rho = 0 # No correlation for clean design
),
seed = 42
)For Bayesian Frameworks
items <- sim_item_params(
n_items = 25,
model = "2pl",
source = "hierarchical",
hierarchical_params = list(
mu = c(0, 0),
tau = c(0.3, 1),
rho = -0.3
),
seed = 42
)References
Glas, C. A. W., & van der Linden, W. J. (2003). Computerized adaptive testing with item cloning. Applied Psychological Measurement, 27(4), 247-261.
Sweeney, S. M., et al. (2022). An investigation of the nature and consequence of the relationship between IRT difficulty and discrimination. Educational Measurement: Issues and Practice, 41(4), 50-67.
Zhang, L., Fellinghauer, C., Geerlings, H., & Sijtsma, K. (2025). Realistic simulation of item difficulties using the Item Response Warehouse. PsyArXiv. https://doi.org/10.31234/osf.io/r5mxv